8IP8
 
 | | Wheat 80S ribosome stalled on AUG-Stop boron dependently | | 分子名称: | 18S ribosomal RNA, 40S ribosomal protein eL8, 40S ribosomal protein eS1, ... | | 著者 | Yokoyama, T, Tanaka, M, Saito, H, Nishimoto, M, Tsuda, K, Sotta, N, Shigematsu, H, Shirouzu, M, Iwasaki, S, Ito, T, Fujiwara, T. | | 登録日 | 2023-03-14 | | 公開日 | 2024-02-21 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Boric acid intercepts 80S ribosome migration from AUG-stop by stabilizing eRF1.
Nat.Chem.Biol., 20, 2024
|
|
8IP9
 
 | | Wheat 40S ribosome in complex with a tRNAi | | 分子名称: | 18S ribosomal RNA, 40S ribosomal protein S23, 40S ribosomal protein eS1, ... | | 著者 | Yokoyama, T, Tanaka, M, Saito, H, Nishimoto, M, Tsuda, K, Sotta, N, Shigematsu, H, Shirouzu, M, Iwasaki, S, Ito, T, Fujiwara, T. | | 登録日 | 2023-03-14 | | 公開日 | 2024-02-21 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Boric acid intercepts 80S ribosome migration from AUG-stop by stabilizing eRF1.
Nat.Chem.Biol., 20, 2024
|
|
8IPB
 
 | | Wheat 80S ribosome pausing on AUG-Stop with cycloheximide | | 分子名称: | 18S ribosomal RNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, 40S ribosomal protein eL8, ... | | 著者 | Yokoyama, T, Tanaka, M, Saito, H, Nishimoto, M, Tsuda, K, Sotta, N, Shigematsu, H, Shirouzu, M, Iwasaki, S, Ito, T, Fujiwara, T. | | 登録日 | 2023-03-14 | | 公開日 | 2024-02-21 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Boric acid intercepts 80S ribosome migration from AUG-stop by stabilizing eRF1.
Nat.Chem.Biol., 20, 2024
|
|
8IPA
 
 | | Wheat 80S ribosome stalled on AUG-Stop boron dependently with cycloheximide | | 分子名称: | 18S ribosomal RNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, 40S ribosomal protein eL8, ... | | 著者 | Yokoyama, T, Tanaka, M, Saito, H, Nishimoto, M, Tsuda, K, Sotta, N, Shigematsu, H, Shirouzu, M, Iwasaki, S, Ito, T, Fujiwara, T. | | 登録日 | 2023-03-14 | | 公開日 | 2024-02-21 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Boric acid intercepts 80S ribosome migration from AUG-stop by stabilizing eRF1.
Nat.Chem.Biol., 20, 2024
|
|
2K2R
 
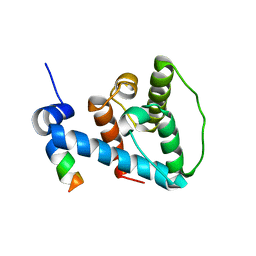 | | The NMR structure of alpha-parvin CH2/paxillin LD1 complex | | 分子名称: | Alpha-parvin, Paxillin | | 著者 | Wang, X, Fukuda, K, Byeon, I, Velyvis, A, Wu, C, Gronenborn, A, Qin, J. | | 登録日 | 2008-04-10 | | 公開日 | 2008-05-27 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Structure of {alpha}-Parvin CH2-Paxillin LD1 Complex Reveals a Novel Modular Recognition for Focal Adhesion Assembly.
J.Biol.Chem., 283, 2008
|
|
2LKO
 
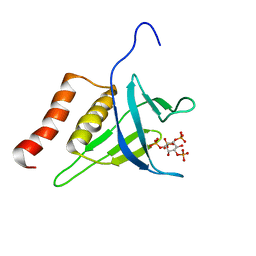 | | Structural Basis of Phosphoinositide Binding to Kindlin-2 Pleckstrin Homology Domain in Regulating Integrin Activation | | 分子名称: | Fermitin family homolog 2, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE | | 著者 | Liu, J, Fukuda, K, Xu, Z. | | 登録日 | 2011-10-17 | | 公開日 | 2011-10-26 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis of phosphoinositide binding to kindlin-2 protein pleckstrin homology domain in regulating integrin activation.
J.Biol.Chem., 286, 2011
|
|
3AY8
 
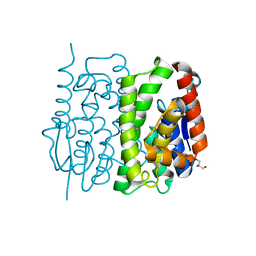 | | Glutathione S-transferase unclassified 2 from Bombyx mori | | 分子名称: | GLYCEROL, Glutathione S-transferase | | 著者 | Kakuta, Y, Usuda, K, Nakashima, T, Kimura, M, Aso, Y, Yamamoto, K. | | 登録日 | 2011-05-02 | | 公開日 | 2011-09-07 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystallographic survey of active sites of an unclassified glutathione transferase from Bombyx mori
Biochim.Biophys.Acta, 1810, 2011
|
|
2ZJV
 
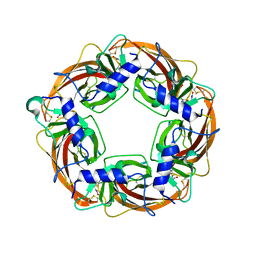 | | Crystal Structure of Lymnaea stagnalis Acetylcholine Binding Protein (Ls-AChBP) Complexed with Clothianidin | | 分子名称: | 1-[(2-chloro-1,3-thiazol-5-yl)methyl]-3-methyl-2-nitroguanidine, Acetylcholine-binding protein | | 著者 | Okajima, T, Ihara, M, Yamashita, A, Oda, T, Morimoto, T, Matsuda, K. | | 登録日 | 2008-03-10 | | 公開日 | 2008-04-08 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structures of Lymnaea stagnalis AChBP in complex with neonicotinoid insecticides imidacloprid and clothianidin
Invert.Neurosci., 8, 2008
|
|
3VK9
 
 | | Crystal structure of delta-class glutathione transferase from silkmoth | | 分子名称: | GLYCEROL, Glutathione S-transferase delta | | 著者 | Kakuta, Y, Usuda, K, Higashiura, A, Suzuki, M, Nakagawa, A, Kimura, M, Yamamoto, K. | | 登録日 | 2011-11-10 | | 公開日 | 2012-10-03 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.001 Å) | | 主引用文献 | Structural basis for catalytic activity of a silkworm Delta-class glutathione transferase
Biochim.Biophys.Acta, 1820, 2012
|
|
5X3Z
 
 | | Solution structure of musashi1 RBD2 in complex with RNA | | 分子名称: | RNA (5'-R(*GP*UP*AP*GP*U)-3'), RNA-binding protein Musashi homolog 1 | | 著者 | Iwaoka, R, Nagata, T, Tsuda, K, Imai, T, Okano, H, Kobayashi, N, Katahira, M. | | 登録日 | 2017-02-09 | | 公開日 | 2017-12-13 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Insight into the Recognition of r(UAG) by Musashi-1 RBD2, and Construction of a Model of Musashi-1 RBD1-2 Bound to the Minimum Target RNA
Molecules, 22, 2017
|
|
5X3Y
 
 | | Refined solution structure of musashi1 RBD2 | | 分子名称: | RNA-binding protein Musashi homolog 1 | | 著者 | Iwaoka, R, Nagata, T, Tsuda, K, Imai, T, Okano, H, Kobayashi, N, Katahira, M. | | 登録日 | 2017-02-09 | | 公開日 | 2017-12-13 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Insight into the Recognition of r(UAG) by Musashi-1 RBD2, and Construction of a Model of Musashi-1 RBD1-2 Bound to the Minimum Target RNA
Molecules, 22, 2017
|
|
2ZJU
 
 | | Crystal Structure of Lymnaea stagnalis Acetylcholine Binding Protein (Ls-AChBP) Complexed with Imidacloprid | | 分子名称: | (2E)-1-[(6-chloropyridin-3-yl)methyl]-N-nitroimidazolidin-2-imine, Acetylcholine-binding protein | | 著者 | Okajima, T, Ihara, M, Yamashita, A, Oda, T, Morimoto, T, Matsuda, K. | | 登録日 | 2008-03-10 | | 公開日 | 2008-04-08 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Crystal structures of Lymnaea stagnalis AChBP in complex with neonicotinoid insecticides imidacloprid and clothianidin
Invert.Neurosci., 8, 2008
|
|
1VAI
 
 | | Structure of e. coli cyclophilin B K163T mutant bound to n-acetyl-ala-ala-pro-ala-7-amino-4-methylcoumarin | | 分子名称: | (ACE)AAPA(MCM), cyclophilin B | | 著者 | Konno, M, Sano, Y, Okudaira, K, Kawaguchi, Y, Yamagishi-Ohmori, Y, Fushinobu, S, Matsuzawa, H. | | 登録日 | 2004-02-17 | | 公開日 | 2004-09-21 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Escherichia coli cyclophilin B binds a highly distorted form of trans-prolyl peptide isomer
Eur.J.Biochem., 271, 2004
|
|
1V9T
 
 | | Structure of E. coli cyclophilin B K163T mutant bound to succinyl-ALA-PRO-ALA-P-nitroanilide | | 分子名称: | (SIN)APA(NIT), cyclophilin B | | 著者 | Konno, M, Sano, Y, Okudaira, K, Kawaguchi, Y, Yamagishi-Ohmori, Y, Fushinobu, S, Matsuzawa, H. | | 登録日 | 2004-02-03 | | 公開日 | 2004-09-21 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Escherichia coli cyclophilin B binds a highly distorted form of trans-prolyl peptide isomer
Eur.J.Biochem., 271, 2004
|
|
1M7W
 
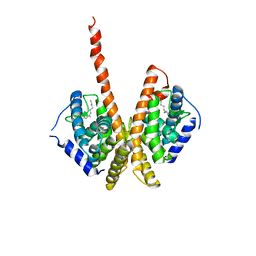 | | HNF4a ligand binding domain with bound fatty acid | | 分子名称: | Hepatocyte nuclear factor 4-alpha, LAURIC ACID | | 著者 | Dhe-Paganon, S, Duda, K, Iwamoto, M, Chi, Y.I, Shoelson, S.E. | | 登録日 | 2002-07-22 | | 公開日 | 2003-07-01 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of the HNF4 alpha ligand binding domain in complex with endogenous fatty acid ligand
J.Biol.Chem., 277, 2002
|
|
1J2A
 
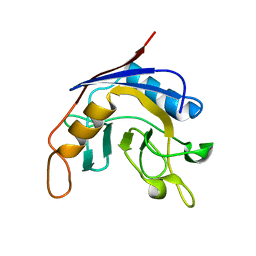 | | Structure of E. coli cyclophilin B K163T mutant | | 分子名称: | cyclophilin B | | 著者 | Konno, M, Sano, Y, Okudaira, K, Kawaguchi, Y, Yamagishi-Ohmori, Y, Fushinobu, S, Matsuzawa, H. | | 登録日 | 2002-12-26 | | 公開日 | 2004-02-10 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Escherichia coli cyclophilin B binds a highly distorted form of trans-prolyl peptide isomer
Eur.J.Biochem., 271, 2004
|
|
1VDN
 
 | |
2DNP
 
 | | Solution structure of RNA binding domain 2 in RNA-binding protein 14 | | 分子名称: | RNA-binding protein 14 | | 著者 | Kusuhara, M, Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2006-04-26 | | 公開日 | 2006-10-26 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of RNA binding domain 2 in RNA-binding protein 14
To be Published
|
|
2DH8
 
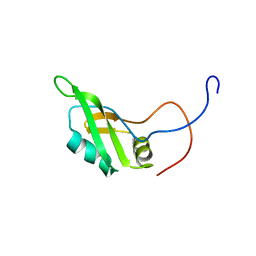 | | Solution structure of the N-terminal RNA binding domain in DAZ-associated protein 1 | | 分子名称: | DAZ-associated protein 1 | | 著者 | Arai, S, Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2006-03-23 | | 公開日 | 2006-09-23 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the N-terminal RNA binding domain in DAZ-associated protein 1
To be Published
|
|
2DH7
 
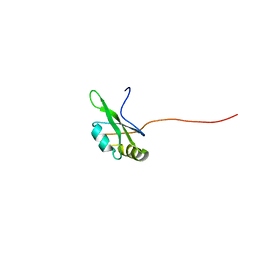 | | Solution structure of the second RNA binding domain in Nucleolysin TIAR | | 分子名称: | Nucleolysin TIAR | | 著者 | Imai, T, Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2006-03-23 | | 公開日 | 2006-09-23 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the second RNA binding domain in Nucleolysin TIAR
To be Published
|
|
2DH9
 
 | | Solution structure of the C-terminal RNA binding domain in Heterogeneous nuclear ribonucleoprotein M | | 分子名称: | Heterogeneous nuclear ribonucleoprotein M | | 著者 | Ochi, T, Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2006-03-23 | | 公開日 | 2006-09-23 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the C-terminal RNA binding domain in Heterogeneous nuclear ribonucleoprotein M
To be Published
|
|
2DHA
 
 | | Solution structure of the second RNA recognition motif in Hypothetical protein FLJ201171 | | 分子名称: | FLJ20171 protein | | 著者 | Imai, T, Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2006-03-23 | | 公開日 | 2006-09-23 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the second RNA recognition motif in Hypothetical protein FLJ201171
To be Published
|
|
2DHG
 
 | | Solution structure of the C-terminal RNA recognition motif in tRNA selenocysteine associated protein | | 分子名称: | tRNA selenocysteine associated protein (SECP43) | | 著者 | Imai, T, Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2006-03-23 | | 公開日 | 2006-09-23 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the C-terminal RNA recognition motif in tRNA selenocysteine associated protein
To be Published
|
|
2E5K
 
 | | Solution structure of SH3 domain in Suppressor of T-cell receptor signaling 1 | | 分子名称: | Suppressor of T-cell receptor signaling 1 | | 著者 | Sukegawa, S, Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2006-12-21 | | 公開日 | 2007-06-26 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of SH3 domain in Suppressor of T-cell receptor signaling 1
To be Published
|
|
2E5H
 
 | | Solution structure of RNA binding domain in Zinc finger CCHC-type and RNA binding motif 1 | | 分子名称: | Zinc finger CCHC-type and RNA-binding motif-containing protein 1 | | 著者 | Iibuchi, H, Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2006-12-21 | | 公開日 | 2007-06-26 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of RNA binding domain in Zinc finger CCHC-type and RNA binding motif 1
To be Published
|
|
