4X6G
 
 | |
5J8C
 
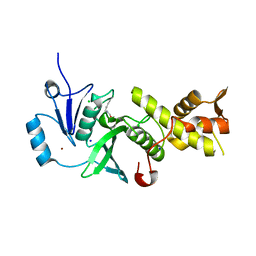 | |
5J8F
 
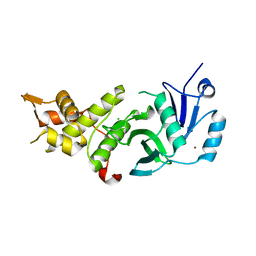 | |
1GUB
 
 | | Hinge-bending motion of D-allose binding protein from Escherichia coli: three open conformations | | 分子名称: | D-ALLOSE-BINDING PERIPLASMIC PROTEIN, NICKEL (II) ION | | 著者 | Magnusson, U, Chaudhuri, B.N, Ko, J, Park, C, Jones, T.A, Mowbray, S.L. | | 登録日 | 2002-01-24 | | 公開日 | 2003-03-06 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structure of D-Allose Binding Protein from Escherichia Coli Bound to D-Allose at 1.8 A Resolution
J.Mol.Biol., 286, 1999
|
|
1GUD
 
 | | Hinge-bending motion of D-allose binding protein from Escherichia coli: three open conformations | | 分子名称: | D-ALLOSE-BINDING PERIPLASMIC PROTEIN, ZINC ION | | 著者 | Magnusson, U, Chaudhuri, B.N, Ko, J, Park, C, Jones, T.A, Mowbray, S.L. | | 登録日 | 2002-01-24 | | 公開日 | 2003-03-06 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Structure of D-Allose Binding Protein from Escherichia Coli Bound to D-Allose at 1.8 A Resolution
J.Mol.Biol., 286, 1999
|
|
5C21
 
 | |
5C22
 
 | | Crystal structure of Zn-bound HlyD from E. coli | | 分子名称: | Chromosomal hemolysin D, ZINC ION | | 著者 | Ha, N.C, Kim, J.S. | | 登録日 | 2015-06-15 | | 公開日 | 2016-02-17 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.302 Å) | | 主引用文献 | Crystal Structure of a Soluble Fragment of the Membrane Fusion Protein HlyD in a Type I Secretion System of Gram-Negative Bacteria
Structure, 24, 2016
|
|
8IV5
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 8H12 and 1C4 (local refinement) | | 分子名称: | Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, heavy chain of 1C4, ... | | 著者 | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | 登録日 | 2023-03-26 | | 公開日 | 2023-08-16 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (3.77 Å) | | 主引用文献 | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8IV8
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 3E2 and 1C4 (local refinement) | | 分子名称: | Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, heavy chain of 1C4, ... | | 著者 | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | 登録日 | 2023-03-26 | | 公開日 | 2023-08-16 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (3.92 Å) | | 主引用文献 | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8IVA
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs XMA01 and 3E2 (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, heavy chain of 3E2, ... | | 著者 | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | 登録日 | 2023-03-26 | | 公開日 | 2023-08-16 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (3.95 Å) | | 主引用文献 | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8IV4
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 8H12 and 3E2 (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, heavy chain of 3E2, ... | | 著者 | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | 登録日 | 2023-03-26 | | 公開日 | 2023-08-16 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (3.59 Å) | | 主引用文献 | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
4GPD
 
 | |
1RPJ
 
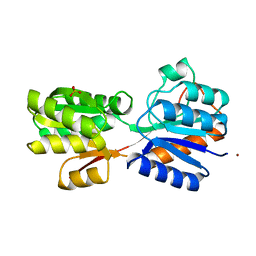 | | CRYSTAL STRUCTURE OF D-ALLOSE BINDING PROTEIN FROM ESCHERICHIA COLI | | 分子名称: | PROTEIN (PRECURSOR OF PERIPLASMIC SUGAR RECEPTOR), SULFATE ION, ZINC ION, ... | | 著者 | Chaudhuri, B, Jones, T.A, Mowbray, S.L. | | 登録日 | 1999-02-04 | | 公開日 | 1999-02-16 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of D-allose binding protein from Escherichia coli bound to D-allose at 1.8 A resolution.
J.Mol.Biol., 286, 1999
|
|
5XVA
 
 | | Crystal Structure of PAK4 in complex with inhibitor CZH216 | | 分子名称: | ETHANOL, Serine/threonine-protein kinase PAK 4, [6-chloranyl-4-[(5-methyl-1H-pyrazol-3-yl)amino]quinazolin-2-yl]-[(3R)-3-methylpiperazin-1-yl]methanone | | 著者 | Zhao, F, Li, H. | | 登録日 | 2017-06-27 | | 公開日 | 2018-02-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.847 Å) | | 主引用文献 | Structure-Based Design of 6-Chloro-4-aminoquinazoline-2-carboxamide Derivatives as Potent and Selective p21-Activated Kinase 4 (PAK4) Inhibitors.
J. Med. Chem., 61, 2018
|
|
5XVG
 
 | | Crystal Structure of PAK4 in complex with inhibitor CZH226 | | 分子名称: | 1,2-ETHANEDIOL, ETHANOL, Serine/threonine-protein kinase PAK 4, ... | | 著者 | Zhao, F, Li, H. | | 登録日 | 2017-06-27 | | 公開日 | 2018-02-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure-Based Design of 6-Chloro-4-aminoquinazoline-2-carboxamide Derivatives as Potent and Selective p21-Activated Kinase 4 (PAK4) Inhibitors.
J. Med. Chem., 61, 2018
|
|
1DXJ
 
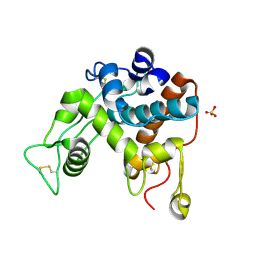 | | Structure of the chitinase from jack bean | | 分子名称: | CLASS II CHITINASE, SULFATE ION | | 著者 | Hahn, M, Hennig, M, Schlesier, B, Hohne, W. | | 登録日 | 2000-01-10 | | 公開日 | 2000-08-29 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of Jack Bean Chitinase
Acta Crystallogr.,Sect.D, 56, 2000
|
|
5XVF
 
 | | Crystal Structure of PAK4 in complex with inhibitor CZH062 | | 分子名称: | 2-(4-azanylpiperidin-1-yl)-6-chloranyl-N-(1-methylimidazol-4-yl)quinazolin-4-amine, Serine/threonine-protein kinase PAK 4 | | 著者 | Zhao, F, Li, H. | | 登録日 | 2017-06-27 | | 公開日 | 2018-02-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.655 Å) | | 主引用文献 | Structure-Based Design of 6-Chloro-4-aminoquinazoline-2-carboxamide Derivatives as Potent and Selective p21-Activated Kinase 4 (PAK4) Inhibitors.
J. Med. Chem., 61, 2018
|
|
6LAT
 
 | |
7E3L
 
 | | Ultrapotent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 58G6 heavy chain, 58G6 light chain, ... | | 著者 | Guo, H, Li, T, Liu, F, Gao, Y, Ji, X, Yang, H. | | 登録日 | 2021-02-09 | | 公開日 | 2021-09-15 | | 最終更新日 | 2022-03-02 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Potent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants.
Nat Commun, 12, 2021
|
|
7E3K
 
 | | Ultrapotent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants | | 分子名称: | 13G9 heavy chain, 13G9 light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Guo, H, Li, T, Liu, F, Gao, Y, Ji, X, Yang, H. | | 登録日 | 2021-02-09 | | 公開日 | 2021-09-15 | | 最終更新日 | 2022-03-02 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Potent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants.
Nat Commun, 12, 2021
|
|
7EAN
 
 | | immune complex of SARS-CoV-2 RBD and cross-neutralizing antibody 6D6 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of SARS-CoV-2 cross-neutralizing mAb 6D6, Light chain of SARS-CoV-2 cross-neutralizing mAb 6D6, ... | | 著者 | Li, T.T, Gu, Y, Li, S.W. | | 登録日 | 2021-03-07 | | 公開日 | 2021-03-31 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Cross-neutralizing antibodies bind a SARS-CoV-2 cryptic site and resist circulating variants.
Nat Commun, 12, 2021
|
|
7WP1
 
 | | Cryo-EM structure of SARS-CoV-2 Mu S6P trimer in complex with neutralizing antibody VacW-209 (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, VacW-209 heavy chain, ... | | 著者 | Zheng, Q, Sun, H, Ju, B, Zhang, Z, Li, S, Xia, N. | | 登録日 | 2022-01-22 | | 公開日 | 2022-07-13 | | 実験手法 | ELECTRON MICROSCOPY (3.77 Å) | | 主引用文献 | Immune escape by SARS-CoV-2 Omicron variant and structural basis of its effective neutralization by a broad neutralizing human antibody VacW-209.
Cell Res., 32, 2022
|
|
7WP5
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron S6P trimer in complex with neutralizing antibody VacW-209 (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spikeprotein S1, VacW-209 heavy chain, ... | | 著者 | Zheng, Q, Sun, H, Ju, B, Zhang, Z, Li, S, Xia, N. | | 登録日 | 2022-01-22 | | 公開日 | 2022-07-13 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Immune escape by SARS-CoV-2 Omicron variant and structural basis of its effective neutralization by a broad neutralizing human antibody VacW-209.
Cell Res., 32, 2022
|
|
7WON
 
 | | Cryo-EM structure of SARS-CoV-2 S2P trimer in complex with neutralizing antibody VacW-209 (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, VacW-209 heavy chain, ... | | 著者 | Zheng, Q, Sun, H, Ju, B, Zhang, Z, Li, S, Xia, N. | | 登録日 | 2022-01-22 | | 公開日 | 2022-07-13 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Immune escape by SARS-CoV-2 Omicron variant and structural basis of its effective neutralization by a broad neutralizing human antibody VacW-209.
Cell Res., 32, 2022
|
|
7WP2
 
 | | Cryo-EM structure of SARS-CoV-2 C.1.2 S6P trimer in complex with neutralizing antibody VacW-209 (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, VacW-209 heavy chain, ... | | 著者 | Zheng, Q, Sun, H, Ju, B, Zhang, Z, Li, S, Xia, N. | | 登録日 | 2022-01-22 | | 公開日 | 2022-07-20 | | 実験手法 | ELECTRON MICROSCOPY (3.52 Å) | | 主引用文献 | Immune escape by SARS-CoV-2 Omicron variant and structural basis of its effective neutralization by a broad neutralizing human antibody VacW-209.
Cell Res., 32, 2022
|
|
