1GEA
 
 | | RECEPTOR-BOUND CONFORMATION OF PACAP21 | | 分子名称: | PITUITARY ADENYLATE CYCLASE ACTIVATING POLYPEPTIDE | | 著者 | Inooka, H, Ohtaki, T, Kitahara, O, Ikegami, T, Endo, S, Kitada, C, Ogi, K, Onda, H, Fujino, M, Shirakawa, M. | | 登録日 | 2000-10-20 | | 公開日 | 2001-04-20 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Conformation of a peptide ligand bound to its G-protein coupled receptor.
Nat.Struct.Biol., 8, 2001
|
|
6I0V
 
 | |
6I0S
 
 | | Crystal structure of DmTailor in complex with UMPNPP | | 分子名称: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, MAGNESIUM ION, Terminal uridylyltransferase Tailor | | 著者 | Kroupova, A, Ivascu, A, Jinek, M. | | 登録日 | 2018-10-26 | | 公開日 | 2018-12-05 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural basis for acceptor RNA substrate selectivity of the 3' terminal uridylyl transferase Tailor.
Nucleic Acids Res., 47, 2019
|
|
6I0T
 
 | |
6I0U
 
 | |
6RW0
 
 | | Crystal structure of ANGEL2, a 2',3'-cyclic phosphatase | | 分子名称: | GLYCEROL, MAGNESIUM ION, POTASSIUM ION, ... | | 著者 | Kroupova, A, Jinek, M. | | 登録日 | 2019-06-03 | | 公開日 | 2020-05-20 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | ANGEL2 is a member of the CCR4 family of deadenylases with 2',3'-cyclic phosphatase activity.
Science, 369, 2020
|
|
6QZK
 
 | | Structure of Clostridium butyricum Argonaute bound to a guide DNA (5' deoxycytidine) and a 19-mer target DNA | | 分子名称: | Clostridium butyricum Argonaute, DNA target (5'-D(T*AP*TP*AP*CP*AP*AP*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*CP*T)-3'), FORMIC ACID, ... | | 著者 | Swarts, D.C, Jinek, M, Hegge, J.W, Van der Oost, J. | | 登録日 | 2019-03-11 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.548 Å) | | 主引用文献 | DNA-guided DNA cleavage at moderate temperatures by Clostridium butyricum Argonaute.
Nucleic Acids Res., 47, 2019
|
|
6RVZ
 
 | | Crystal structure of ANGEL2, a 2',3'-cyclic phosphatase, in complex with adenosine-2',3'-vanadate | | 分子名称: | ADENOSINE, MAGNESIUM ION, POTASSIUM ION, ... | | 著者 | Kroupova, A, Jinek, M. | | 登録日 | 2019-06-03 | | 公開日 | 2020-05-20 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | ANGEL2 is a member of the CCR4 family of deadenylases with 2',3'-cyclic phosphatase activity.
Science, 369, 2020
|
|
5AF2
 
 | |
3ICQ
 
 | | Karyopherin nuclear state | | 分子名称: | Exportin-T, GTP-binding nuclear protein GSP1/CNR1, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Cook, A.G, Fukuhara, N, Jinek, M, Conti, E. | | 登録日 | 2009-07-18 | | 公開日 | 2009-08-25 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structures of the tRNA export factor in the nuclear and cytosolic states
Nature, 461, 2009
|
|
3IBV
 
 | | Karyopherin cytosolic state | | 分子名称: | CALCIUM ION, Exportin-T | | 著者 | Cook, A.G, Fukuhara, N, Jinek, M, Conti, E. | | 登録日 | 2009-07-17 | | 公開日 | 2009-08-25 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structures of the tRNA export factor in the nuclear and cytosolic states
Nature, 461, 2009
|
|
1WS8
 
 | | Crystal Structure of Mavicyanin from Cucurbita pepo medullosa (Zucchini) | | 分子名称: | COPPER (II) ION, GLYCEROL, mavicyanin | | 著者 | Xie, Y, Inoue, T, Miyamoto, Y, Matsumura, H, Kunishige, K, Yamaguchi, K, Nojini, M, Suzuki, S, Kai, Y. | | 登録日 | 2004-11-02 | | 公開日 | 2004-11-23 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural reorganization of the copper binding site involving Thr15 of mavicyanin from Cucurbita pepo medullosa (zucchini) upon reduction.
J.Biochem.(Tokyo), 137, 2005
|
|
2Y8Y
 
 | |
2Y8W
 
 | |
2Y9H
 
 | |
5FW2
 
 | | Crystal structure of SpCas9 variant EQR bound to sgRNA and TGAG PAM target DNA | | 分子名称: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MAGNESIUM ION, NON-TARGET DNA STRAND, ... | | 著者 | Anders, C, Bargsten, K, Jinek, M. | | 登録日 | 2016-02-11 | | 公開日 | 2016-06-15 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.676 Å) | | 主引用文献 | Structural Plasticity of Pam Recognition by Engineered Variants of the RNA-Guided Endonuclease Cas9.
Mol.Cell, 61, 2016
|
|
5NG6
 
 | |
5FW1
 
 | | Crystal structure of SpyCas9 variant VQR bound to sgRNA and TGAG PAM target DNA | | 分子名称: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MAGNESIUM ION, NON-TARGET DNA STRAND, ... | | 著者 | Anders, C, Bargsten, K, Jinek, M. | | 登録日 | 2016-02-11 | | 公開日 | 2016-04-20 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.499 Å) | | 主引用文献 | Structural Plasticity of Pam Recognition by Engineered Variants of the RNA-Guided Endonuclease Cas9.
Mol.Cell, 61, 2016
|
|
1WS7
 
 | | Crystal Structure of Mavicyanin from Cucurbita pepo medullosa (Zucchini) | | 分子名称: | COPPER (I) ION, Mavicyanin | | 著者 | Xie, Y, Inoue, T, Miyamoto, Y, Matsumura, H, Kataoka, K, Yamaguchi, K, Nojini, M, Suzuki, S, Kai, Y. | | 登録日 | 2004-11-02 | | 公開日 | 2004-11-23 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural reorganization of the copper binding site involving Thr15 of mavicyanin from Cucurbita pepo medullosa (zucchini) upon reduction.
J.Biochem.(Tokyo), 137, 2005
|
|
5NFV
 
 | | Crystal structure of catalytically inactive FnCas12 mutant bound to an R-loop structure containing a pre-crRNA mimic and full-length DNA target | | 分子名称: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CRISPR-associated endonuclease Cpf1, DNA non-target strand, ... | | 著者 | Swarts, D.C, van der Oost, J, Jinek, M. | | 登録日 | 2017-03-16 | | 公開日 | 2017-06-14 | | 実験手法 | X-RAY DIFFRACTION (2.501 Å) | | 主引用文献 | Structural Basis for Guide RNA Processing and Seed-Dependent DNA Targeting by CRISPR-Cas12a.
Mol. Cell, 66, 2017
|
|
2XLI
 
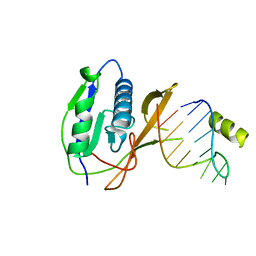 | | Crystal structure of the Csy4-crRNA complex, monoclinic form | | 分子名称: | 5'-R(*CP*UP*GP*CP*CP*GP*UP*AP*UP*AP*GP*GP*CP*A*DG*C)-3', CSY4 ENDORIBONUCLEASE | | 著者 | Haurwitz, R.E, Jinek, M, Wiedenheft, B, Zhou, K, Doudna, J.A. | | 登録日 | 2010-07-20 | | 公開日 | 2010-09-22 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | Sequence- and Structure-Specific RNA Processing by a Crispr Endonuclease.
Science, 329, 2010
|
|
5FSH
 
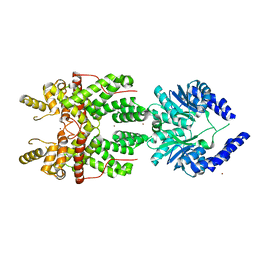 | |
2XLJ
 
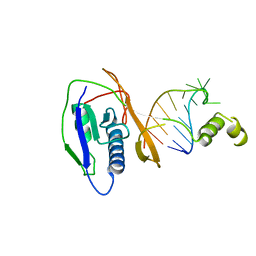 | | Crystal structure of the Csy4-crRNA complex, hexagonal form | | 分子名称: | 5'-R(*CP*UP*GP*CP*CP*GP*UP*AP*UP*AP*GP*GP*CP*A*DG*C)-3', CSY4 ENDORIBONUCLEASE | | 著者 | Haurwitz, R.E, Jinek, M, Wiedenheft, B, Zhou, K, Doudna, J.A. | | 登録日 | 2010-07-20 | | 公開日 | 2010-09-22 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Sequence- and Structure-Specific RNA Processing by a Crispr Endonuclease.
Science, 329, 2010
|
|
2XLK
 
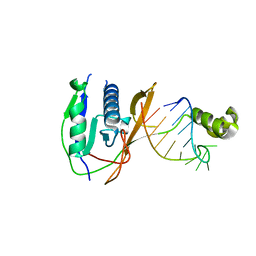 | | Crystal structure of the Csy4-crRNA complex, orthorhombic form | | 分子名称: | 5'-R(*CP*UP*GP*CP*CP*GP*UP*AP*UP*AP*GP*GP*CP*A*DG*C)-3', CSY4 ENDORIBONUCLEASE | | 著者 | Haurwitz, R.E, Jinek, M, Wiedenheft, B, Zhou, K, Doudna, J.A. | | 登録日 | 2010-07-21 | | 公開日 | 2010-09-22 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.805 Å) | | 主引用文献 | Sequence- and Structure-Specific RNA Processing by a Crispr Endonuclease.
Science, 329, 2010
|
|
5FW3
 
 | | Crystal structure of SpCas9 variant VRER bound to sgRNA and TGCG PAM target DNA | | 分子名称: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MAGNESIUM ION, NON-TARGET DNA STRAND, ... | | 著者 | Anders, C, Bargsten, K, Jinek, M. | | 登録日 | 2016-02-11 | | 公開日 | 2016-06-15 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural Plasticity of Pam Recognition by Engineered Variants of the RNA-Guided Endonuclease Cas9.
Mol.Cell, 61, 2016
|
|
