1MXP
 
 | | Solution structure of the ribbon disulfide bond isomer of alpha-conotoxin AuIB | | 分子名称: | alpha-conotoxin AuIB | | 著者 | Dutton, J.L, Bansal, P.S, Hogg, R.C, Adams, D.J, Alewood, P.F, Craik, D.J. | | 登録日 | 2002-10-03 | | 公開日 | 2002-12-30 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A New Level of Conotoxin Diversity, a Non-native Disulfide Bond Connectivity in alpha -Conotoxin AuIB Reduces Structural Definition but Increases Biological Activity.
J.Biol.Chem., 277, 2002
|
|
1MS6
 
 | | Dipeptide Nitrile Inhibitor Bound to Cathepsin S. | | 分子名称: | Cathepsin S, MORPHOLINE-4-CARBOXYLIC ACID [1S-(2-BENZYLOXY-1R-CYANO-ETHYLCARBAMOYL)-3-METHYL-BUTYL]AMIDE | | 著者 | Ward, Y.D, Thomson, D.S, Frye, L.L, Cywin, C.L, Morwick, T, Emmanuel, M.J, Zindell, R, McNeil, D, Bekkali, Y, Giradot, M, Hrapchak, M, DeTuri, M, Crane, K, White, D, Pav, S, Wang, Y, Hao, M.H, Grygon, C.A, Labadia, M.E, Freeman, D.M, Davidson, W, Hopkins, J.L, Brown, M.L, Spero, D.M. | | 登録日 | 2002-09-19 | | 公開日 | 2003-04-22 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Design and synthesis of dipeptide nitriles as reversible and potent Cathepsin S inhibitors
J.Med.Chem., 45, 2002
|
|
1MXS
 
 | | Crystal structure of 2-keto-3-deoxy-6-phosphogluconate (KDPG) aldolase from Pseudomonas putida. | | 分子名称: | KDPG Aldolase, SULFATE ION | | 著者 | Watanabe, L, Bell, B.J, Lebioda, L, Rios-Steiner, J.L, Tulinsky, A, Arni, R.K. | | 登録日 | 2002-10-03 | | 公開日 | 2003-09-16 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of 2-keto-3-deoxy-6-phosphogluconate (KDPG) aldolase from Pseudomonas putida.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1MXN
 
 | | Solution structure of alpha-conotoxin AuIB | | 分子名称: | alpha-conotoxin AuIB | | 著者 | Dutton, J.L, Bansal, P.S, Hogg, R.C, Adams, D.J, Alewood, P.F, Craik, D.J. | | 登録日 | 2002-10-02 | | 公開日 | 2002-12-30 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A New Level of Conotoxin Diversity, a Non-native Disulfide Bond Connectivity in alpha -Conotoxin AuIB Reduces Structural Definition but Increases Biological Activity.
J.Biol.Chem., 277, 2002
|
|
1HSB
 
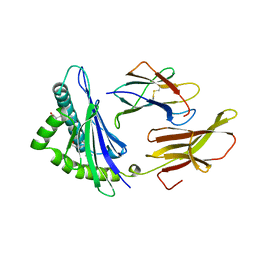 | | DIFFERENT LENGTH PEPTIDES BIND TO HLA-AW68 SIMILARLY AT THEIR ENDS BUT BULGE OUT IN THE MIDDLE | | 分子名称: | ALANINE, ARGININE, BETA 2-MICROGLOBULIN, ... | | 著者 | Guo, H.-C, Strominger, J.L, Wiley, D.C. | | 登録日 | 1993-03-30 | | 公開日 | 1993-10-31 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Different length peptides bind to HLA-Aw68 similarly at their ends but bulge out in the middle.
Nature, 360, 1992
|
|
5K5P
 
 | |
3TPJ
 
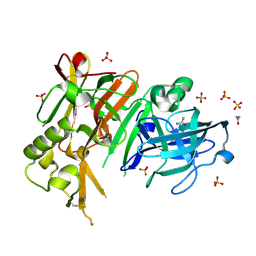 | | APO structure of BACE1 | | 分子名称: | Beta-secretase 1, CHLORIDE ION, SULFATE ION, ... | | 著者 | Xu, Y.C, Li, M.J, Greenblatt, H, Chen, T.T, Silman, I, Sussman, J.L. | | 登録日 | 2011-09-08 | | 公開日 | 2011-11-23 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | Flexibility of the flap in the active site of BACE1 as revealed by crystal structures and molecular dynamics simulations
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3TP5
 
 | |
3TPL
 
 | | APO Structure of BACE1 | | 分子名称: | Beta-secretase 1, CHLORIDE ION, SULFATE ION | | 著者 | Xu, Y.C, Li, M.J, Greenblatt, H, Chen, T.T, Silman, I, Sussman, J.L. | | 登録日 | 2011-09-08 | | 公開日 | 2011-11-23 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Flexibility of the flap in the active site of BACE1 as revealed by crystal structures and molecular dynamics simulations
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3TU3
 
 | | 1.92 Angstrom resolution crystal structure of the full-length SpcU in complex with full-length ExoU from the type III secretion system of Pseudomonas aeruginosa | | 分子名称: | ExoU, ExoU chaperone | | 著者 | Halavaty, A.S, Borek, D, Otwinowski, Z, Minasov, G, Veesenmeyer, J.L, Tyson, G, Shuvalova, L, Hauser, A.R, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-09-15 | | 公開日 | 2012-05-23 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Structure of the Type III Secretion Effector Protein ExoU in Complex with Its Chaperone SpcU.
Plos One, 7, 2012
|
|
3TPR
 
 | | Crystal structure of BACE1 complexed with an inhibitor | | 分子名称: | Beta-secretase 1, CHLORIDE ION, N-[(1S,2R)-1-BENZYL-3-(CYCLOPROPYLAMINO)-2-HYDROXYPROPYL]-5-[METHYL(METHYLSULFONYL)AMINO]-N'-[(1R)-1-PHENYLETHYL]ISOPHTHALAMIDE | | 著者 | Xu, Y.C, Li, M.J, Greenblatt, H, Chen, T.T, Silman, I, Sussman, J.L. | | 登録日 | 2011-09-08 | | 公開日 | 2011-11-23 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Flexibility of the flap in the active site of BACE1 as revealed by crystal structures and molecular dynamics simulations
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3TZ4
 
 | | Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/S-alpha-chloroisocaproate complex with ADP | | 分子名称: | (2S)-2-chloro-4-methylpentanoic acid, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Tso, S.C, Chuang, J.L, Gui, W.J, Wynn, R.M, Li, J, Chuang, D.T. | | 登録日 | 2011-09-27 | | 公開日 | 2012-10-03 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structure-based design and mechanisms of allosteric inhibitors for mitochondrial branched-chain alpha-ketoacid dehydrogenase kinase.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3TP8
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS L36E at cryogenic temperature | | 分子名称: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | 著者 | Robinson, A.C, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | 登録日 | 2011-09-07 | | 公開日 | 2011-09-21 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of Staphylococcal nuclease variant Delta+PHS L36E at cryogenic temperature
To be Published
|
|
3TWE
 
 | | Crystal Structure of the de novo designed peptide alpha4H | | 分子名称: | ACETYL GROUP, TRIETHYLENE GLYCOL, alpha4H | | 著者 | Buer, B.C, Meagher, J.L, Stuckey, J.A, Marsh, E.N.G. | | 登録日 | 2011-09-21 | | 公開日 | 2012-03-14 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | Structural basis for the enhanced stability of highly fluorinated proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3TWG
 
 | |
3TWF
 
 | | Crystal structure of the de novo designed fluorinated peptide alpha4F3a | | 分子名称: | ACETYL GROUP, SODIUM ION, TRIETHYLENE GLYCOL, ... | | 著者 | Buer, B.C, Meagher, J.L, Stuckey, J.A, Marsh, E.N.G. | | 登録日 | 2011-09-21 | | 公開日 | 2012-03-14 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Structural basis for the enhanced stability of highly fluorinated proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UFM
 
 | |
3UDG
 
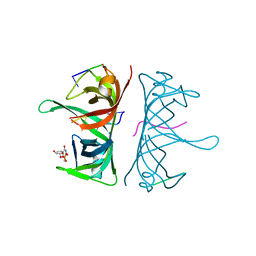 | | Structure of Deinococcus radiodurans SSB bound to ssDNA | | 分子名称: | 5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3', Single-stranded DNA-binding protein, THYMIDINE-5'-PHOSPHATE | | 著者 | George, N.P, Ngo, K.V, Chitteni-Patu, S, Norais, C.A, Battista, J.R, Cox, M.M, Keck, J.L. | | 登録日 | 2011-10-28 | | 公開日 | 2012-05-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure and Cellular Dynamics of Deinococcus radiodurans Single-stranded DNA (ssDNA)-binding Protein (SSB)-DNA Complexes.
J.Biol.Chem., 287, 2012
|
|
3UF7
 
 | |
2AAS
 
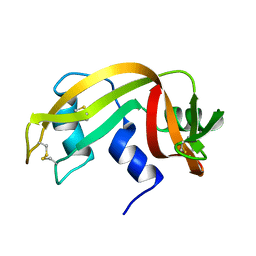 | | HIGH-RESOLUTION THREE-DIMENSIONAL STRUCTURE OF RIBONUCLEASE A IN SOLUTION BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | 分子名称: | RIBONUCLEASE A | | 著者 | Santoro, J, Gonzalez, C, Bruix, M, Neira, J.L, Nieto, J.L, Herranz, J, Rico, M. | | 登録日 | 1992-11-20 | | 公開日 | 1994-01-31 | | 最終更新日 | 2017-11-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | High-resolution three-dimensional structure of ribonuclease A in solution by nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 229, 1993
|
|
2WWD
 
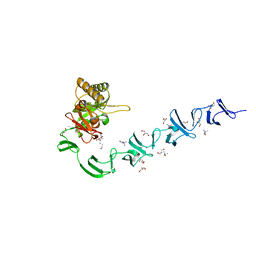 | | 3D-structure of the modular autolysin LytC from Streptococcus pneumoniae in complex with pneummococcal peptidoglycan fragment | | 分子名称: | 1,4-BETA-N-ACETYLMURAMIDASE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-alpha-muramic acid, ALANINE, ... | | 著者 | Perez-Dorado, I, Sanles, R, Hermoso, J.A, Gonzalez, A, Garcia, A, Garcia, P, Garcia, J.L. | | 登録日 | 2009-10-22 | | 公開日 | 2010-04-21 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Insights Into Pneumococcal Fratricide from the Crystal Structures of the Modular Killing Factor Lytc.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2WW5
 
 | | 3D-structure of the modular autolysin LytC from Streptococcus pneumoniae at 1.6 A resolution | | 分子名称: | 1,4-BETA-N-ACETYLMURAMIDASE, CHLORIDE ION, CHOLINE ION, ... | | 著者 | Perez-Dorado, I, Sanles, R, Hermoso, J.A, Gonzalez, A, Garcia, A, Garcia, P, Garcia, J.L, Menendez, M. | | 登録日 | 2009-10-21 | | 公開日 | 2010-04-21 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | Insights Into Pneumococcal Fratricide from the Crystal Structures of the Modular Killing Factor Lytc.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2WWC
 
 | | 3D-structure of the modular autolysin LytC from Streptococcus pneumoniae in complex with synthetic peptidoglycan ligand | | 分子名称: | 1,4-BETA-N-ACETYLMURAMIDASE, CHOLINE ION, GLYCEROL | | 著者 | Perez-Dorado, I, Sanles, R, Hermoso, J.A, Gonzalez, A, Garcia, A, Garcia, P, Garcia, J.L. | | 登録日 | 2009-10-22 | | 公開日 | 2010-04-21 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Insights Into Pneumococcal Fratricide from the Crystal Structures of the Modular Killing Factor Lytc.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KPY
 
 | | Crystal Structure of hPNMT in Complex AdoHcy and 6-Chlorooxindole | | 分子名称: | 6-chloro-1,3-dihydro-2H-indol-2-one, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Drinkwater, N, Martin, J.L. | | 登録日 | 2009-11-17 | | 公開日 | 2010-09-29 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Fragment-based screening by X-ray crystallography, MS and isothermal titration calorimetry to identify PNMT (phenylethanolamine N-methyltransferase) inhibitors.
Biochem.J., 431, 2010
|
|
6WSH
 
 | |
