6DVK
 
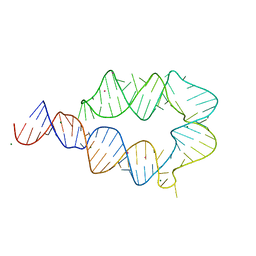 | | Computationally designed mini tetraloop-tetraloop receptor by the RNAMake program - construct 6 (miniTTR 6) | | 分子名称: | COBALT (II) ION, MAGNESIUM ION, RNA (95-MER) | | 著者 | Eiler, D.R, Yesselman, J.D, Costantino, D.A, Das, R, Kieft, J.S. | | 登録日 | 2018-06-24 | | 公開日 | 2019-06-26 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Computational design of three-dimensional RNA structure and function.
Nat Nanotechnol, 14, 2019
|
|
3FZW
 
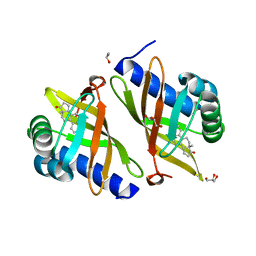 | | Crystal Structure of Ketosteroid Isomerase D40N-D103N from Pseudomonas putida (pKSI) with bound equilenin | | 分子名称: | EQUILENIN, GLYCEROL, ISOPROPYL ALCOHOL, ... | | 著者 | Caaveiro, J.M.M, Ringe, D, Petsko, G.A. | | 登録日 | 2009-01-26 | | 公開日 | 2009-06-02 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | Hydrogen bond coupling in the ketosteroid isomerase active site.
Biochemistry, 48, 2009
|
|
3CPO
 
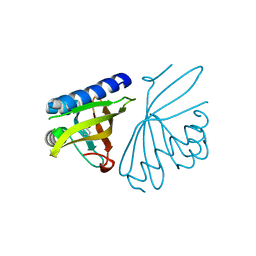 | | Crystal structure of ketosteroid isomerase D40N with bound 2-fluorophenol | | 分子名称: | 2-fluorophenol, Delta(5)-3-ketosteroid isomerase | | 著者 | Caaveiro, J.M.M, Pybus, B, Ringe, D, Petsko, G. | | 登録日 | 2008-03-31 | | 公開日 | 2008-09-23 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.24 Å) | | 主引用文献 | Testing geometrical discrimination within an enzyme active site: constrained hydrogen bonding in the ketosteroid isomerase oxyanion hole
J.Am.Chem.Soc., 130, 2008
|
|
1B4S
 
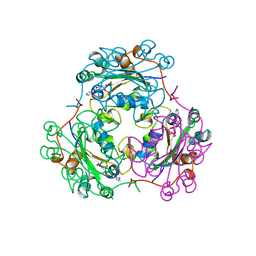 | |
3VGN
 
 | | Crystal Structure of Ketosteroid Isomerase D40N from Pseudomonas putida (pKSI) with bound 3-fluoro-4-nitrophenol | | 分子名称: | 3-fluoro-4-nitrophenol, Steroid Delta-isomerase | | 著者 | Caaveiro, J.M.M, Pybus, B, Ringe, D, Petsko, G.A, Sigala, P.A. | | 登録日 | 2011-08-16 | | 公開日 | 2012-08-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Quantitative dissection of hydrogen bond-mediated proton transfer in the ketosteroid isomerase active site
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2PZV
 
 | | Crystal Structure of Ketosteroid Isomerase D40N from Pseudomonas Putida (pksi) with bound Phenol | | 分子名称: | PHENOL, Steroid Delta-isomerase | | 著者 | Pybus, B, Caaveiro, J.M.M, Petsko, G.A, Ringe, D. | | 登録日 | 2007-05-18 | | 公開日 | 2007-06-12 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Testing Electrostatic complementarity in Enzyme Catalysis: Hydrogen Bonding in the Ketosteroid Isomerase Oxyanion Hole
PLoS Biol., 4, 2006
|
|
7JU1
 
 | | The FARFAR-NMR Ensemble of 29-mer HIV-1 Trans-activation Response Element RNA (N=20) | | 分子名称: | RNA (29-MER) | | 著者 | Shi, H, Rangadurai, A, Roy, R, Yesselman, J.D, Al-Hashimi, H.M. | | 登録日 | 2020-08-18 | | 公開日 | 2020-10-07 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Rapid and accurate determination of atomistic RNA dynamic ensemble models using NMR and structure prediction
Nat Commun, 11, 2020
|
|
2RH6
 
 | |
8EZO
 
 | | Lysozyme Anomalous Dataset at 220 K and 7.1 keV | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | 著者 | Doukov, T, Yabukarski, F. | | 登録日 | 2022-11-01 | | 公開日 | 2023-03-15 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Obtaining anomalous and ensemble information from protein crystals from 220 K up to physiological temperatures.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
3O6Y
 
 | | Robust computational design, optimization, and structural characterization of retroaldol enzymes | | 分子名称: | Retro-Aldolase, SULFATE ION | | 著者 | Althoff, E.A, Wang, L, Jiang, L, Moody, J, Bolduc, J, Lassila, J.K, Wang, Z.Z, Smith, M, Hari, S, Herschlag, D, Stoddard, B.L, Baker, D. | | 登録日 | 2010-07-29 | | 公開日 | 2011-06-29 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.091 Å) | | 主引用文献 | Structural analyses of covalent enzyme-substrate analog complexes reveal strengths and limitations of de novo enzyme design.
J.Mol.Biol., 415, 2012
|
|
3NXF
 
 | | Robust computational design, optimization, and structural characterization of retroaldol enzymes | | 分子名称: | Retro-Aldolase, SULFATE ION | | 著者 | Althoff, E.A, Jiang, L, Wang, L, Lassila, J.K, Moody, J, Bolduc, J, Wang, Z.Z, Smith, M, Hari, S, Herschlag, D, Stoddard, B.L, Baker, D. | | 登録日 | 2010-07-13 | | 公開日 | 2011-06-29 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural analyses of covalent enzyme-substrate analog complexes reveal strengths and limitations of de novo enzyme design.
J.Mol.Biol., 415, 2012
|
|
