1XP4
 
 | | Crystal structure of a peptidoglycan synthesis regulatory factor (PBP3) from Streptococcus pneumoniae | | 分子名称: | D-alanyl-D-alanine carboxypeptidase, IODIDE ION, SULFATE ION | | 著者 | Morlot, C, Pernot, L, Le Gouellec, A, Di Guilmi, A.M, Vernet, T, Dideberg, O, Dessen, A. | | 登録日 | 2004-10-08 | | 公開日 | 2004-11-09 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of a peptidoglycan synthesis regulatory factor (PBP3) from Streptococcus pneumoniae
J.Biol.Chem., 280, 2005
|
|
2AAF
 
 | |
2A9G
 
 | | Structure of C406A arginine deiminase in complex with L-arginine | | 分子名称: | ARGININE, Arginine deiminase | | 著者 | Galkin, A, Lu, X, Dunaway-Mariano, D, Herzberg, O. | | 登録日 | 2005-07-11 | | 公開日 | 2005-08-09 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal Structures Representing the Michaelis Complex and the Thiouronium Reaction Intermediate of Pseudomonas aeruginosa Arginine Deiminase.
J.Biol.Chem., 280, 2005
|
|
2ACI
 
 | |
1MOV
 
 | | Crystal structure of Coral protein mutant | | 分子名称: | GFP-like non-fluorescent chromoprotein, IODIDE ION | | 著者 | Prescott, M, Ling, M, Beddoe, T, Oakley, A.J, Dove, S, Hoegh-Guldberg, O, Devenish, R.J, Rossjohn, J. | | 登録日 | 2002-09-10 | | 公開日 | 2003-04-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The 2.2 a crystal structure of a pocilloporin pigment reveals a nonplanar chromophore conformation.
Structure, 11, 2003
|
|
8SOT
 
 | |
1H80
 
 | | 1,3-ALPHA-1,4-BETA-D-GALACTOSE-4-SULFATE- 3,6-ANHYDRO-D-GALACTOSE-2-SULFATE 4 GALACTOHYDROLASE | | 分子名称: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | 著者 | Michel, G, Chantalat, L, Dideberg, O. | | 登録日 | 2001-01-22 | | 公開日 | 2001-11-27 | | 最終更新日 | 2018-10-24 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The Iota-Carrageenase of Alteromonas Fortis. A Beta-Helix Fold-Containing Enzyme for the Degradation of a Highly Polyanionic Polysaccharide
J.Biol.Chem., 276, 2001
|
|
1GAE
 
 | | COMPARISON OF THE STRUCTURES OF WILD TYPE AND A N313T MUTANT OF ESCHERICHIA COLI GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASES: IMPLICATION FOR NAD BINDING AND COOPERATIVITY | | 分子名称: | D-GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Duee, E, Olivier-Deyris, L, Fanchon, E, Corbier, C, Branlant, G, Dideberg, O. | | 登録日 | 1995-10-24 | | 公開日 | 1996-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Comparison of the structures of wild-type and a N313T mutant of Escherichia coli glyceraldehyde 3-phosphate dehydrogenases: implication for NAD binding and cooperativity.
J.Mol.Biol., 257, 1996
|
|
1GGO
 
 | | T453A MUTANT OF PYRUVATE, PHOSPHATE DIKINASE | | 分子名称: | PROTEIN (PYRUVATE, PHOSPHATE DIKINASE), SULFATE ION | | 著者 | Li, Z, Herzberg, O. | | 登録日 | 2000-08-29 | | 公開日 | 2001-01-10 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Identification of domain-domain docking sites within Clostridium symbiosum pyruvate phosphate dikinase by amino acid replacement.
J.Biol.Chem., 275, 2000
|
|
1GAD
 
 | | COMPARISON OF THE STRUCTURES OF WILD TYPE AND A N313T MUTANT OF ESCHERICHIA COLI GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASES: IMPLICATION FOR NAD BINDING AND COOPERATIVITY | | 分子名称: | D-GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Duee, E, Olivier-Deyris, L, Fanchon, E, Corbier, C, Branlant, G, Dideberg, O. | | 登録日 | 1995-10-24 | | 公開日 | 1996-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Comparison of the structures of wild-type and a N313T mutant of Escherichia coli glyceraldehyde 3-phosphate dehydrogenases: implication for NAD binding and cooperativity.
J.Mol.Biol., 257, 1996
|
|
3ZNB
 
 | | METALLO-BETA-LACTAMASE (ZN, HG-BOUND FORM) | | 分子名称: | MERCURY (II) ION, METALLO-BETA-LACTAMASE, SODIUM ION, ... | | 著者 | Concha, N.O, Herzberg, O. | | 登録日 | 1997-10-15 | | 公開日 | 1998-01-28 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structures of the cadmium- and mercury-substituted metallo-beta-lactamase from Bacteroides fragilis.
Protein Sci., 6, 1997
|
|
7RFV
 
 | | Tailspike protein 4 (TSP4) from phage CBA120, residues 1-250, obtained in the presence of PEG8000 | | 分子名称: | Tailspike protein | | 著者 | Chao, K, Shang, X, Grenfield, J, Linden, S.B, Nelson, D.C, Herzberg, O. | | 登録日 | 2021-07-14 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structure of Escherichia coli O157:H7 bacteriophage CBA120 tailspike protein 4 baseplate anchor and tailspike assembly domains (TSP4-N).
Sci Rep, 12, 2022
|
|
7RFO
 
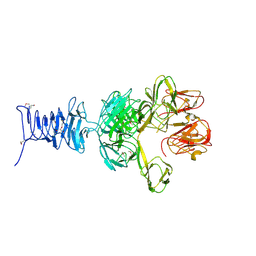 | | SeMet Tailspike protein 4 (TSP4) phage CBA120, residues 1-335, obtained in the presence of LiSO4 | | 分子名称: | Tailspike protein | | 著者 | Chao, K, Shang, X, Grenfield, J, Linden, S.B, Nelson, D.C, Herzberg, O. | | 登録日 | 2021-07-14 | | 公開日 | 2022-02-23 | | 実験手法 | X-RAY DIFFRACTION (3.02 Å) | | 主引用文献 | Structure of Escherichia coli O157:H7 bacteriophage CBA120 tailspike protein 4 baseplate anchor and tailspike assembly domains (TSP4-N).
Sci Rep, 12, 2022
|
|
7REJ
 
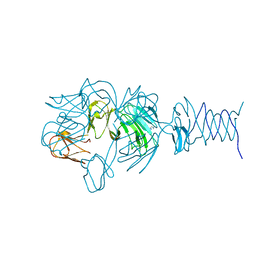 | | Tailspike protein 4 (TSP4) from phage CBA120, residues 1-335, obtained in the presence of NaK-Tartrate | | 分子名称: | IMIDAZOLE, Tailspike protein | | 著者 | Chao, K, Shang, X, Grenfield, J, Linden, S.B, Nelson, D.C, Herzberg, O. | | 登録日 | 2021-07-13 | | 公開日 | 2022-02-23 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure of Escherichia coli O157:H7 bacteriophage CBA120 tailspike protein 4 baseplate anchor and tailspike assembly domains (TSP4-N).
Sci Rep, 12, 2022
|
|
2XD5
 
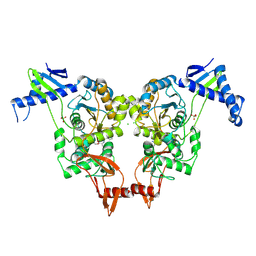 | | Structural insights into the catalytic mechanism and the role of Streptococcus pneumoniae PBP1b | | 分子名称: | CHLORIDE ION, N-BENZOYL-D-ALANINE, PENICILLIN-BINDING PROTEIN 1B, ... | | 著者 | Macheboeuf, P, Lemaire, D, Jamin, M, Dideberg, O, Dessen, A. | | 登録日 | 2010-04-29 | | 公開日 | 2010-05-26 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural Insights Into the Catalytic Mechanism and the Role of Streptococcus Pneumoniae Pbp1B
To be Published
|
|
1ORT
 
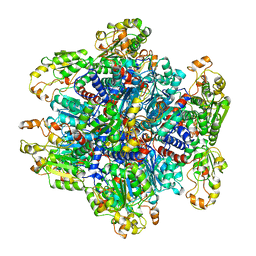 | | ORNITHINE TRANSCARBAMOYLASE FROM PSEUDOMONAS AERUGINOSA | | 分子名称: | ORNITHINE TRANSCARBAMOYLASE | | 著者 | Villeret, V, Dideberg, O. | | 登録日 | 1995-08-24 | | 公開日 | 1996-12-07 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structure of Pseudomonas aeruginosa catabolic ornithine transcarbamoylase at 3.0-A resolution: a different oligomeric organization in the transcarbamoylase family.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1OQF
 
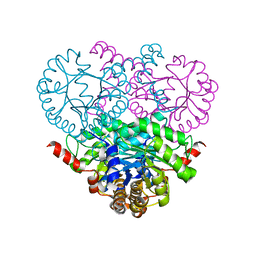 | | Crystal structure of the 2-methylisocitrate lyase | | 分子名称: | 2-methylisocitrate lyase | | 著者 | Liu, S, Lu, Z, Dunaway-Mariano, D, Herzberg, O, Structure 2 Function Project (S2F) | | 登録日 | 2003-03-08 | | 公開日 | 2004-04-27 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Crystal structures of 2-methylisocitrate lyase in complex with product and with isocitrate inhibitor provide insight into lyase substrate specificity, catalysis and evolution.
Biochemistry, 44, 2005
|
|
1NNX
 
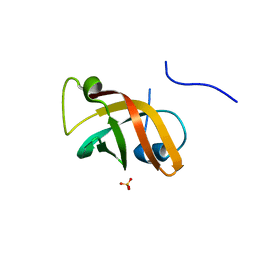 | | Structure of the hypothetical protein ygiW from E. coli. | | 分子名称: | Protein ygiW, SULFATE ION | | 著者 | Lehmann, C, Galkin, A, Pullalarevu, S, Sarikaya, E, Krajewski, W, Lim, K, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | 登録日 | 2003-01-14 | | 公開日 | 2004-03-09 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structure of the hypothetical protein ygiW from E. coli.
To be Published
|
|
3KZF
 
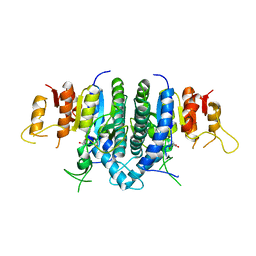 | | Structure of Giardia Carbamate Kinase | | 分子名称: | Carbamate kinase, GLYCEROL | | 著者 | Galkin, A, Herzberg, O. | | 登録日 | 2009-12-08 | | 公開日 | 2010-04-07 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | X-ray structure and characterization of carbamate kinase from the human parasite Giardia lamblia.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
1NO5
 
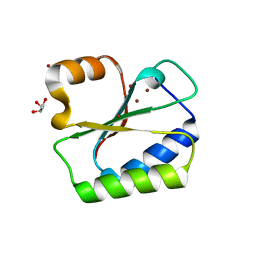 | | Structure of HI0073 from Haemophilus influenzae, the nucleotide binding domain of the HI0073/HI0074 two protein nucleotidyl transferase. | | 分子名称: | GLYCEROL, Hypothetical protein HI0073, SODIUM ION, ... | | 著者 | Lehmann, C, Pullalarevu, S, Galkin, A, Krajewski, W, Willis, M.A, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | 登録日 | 2003-01-15 | | 公開日 | 2004-03-16 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of HI0073 from Haemophilus influenzae, the nucleotide-binding domain of a two-protein nucleotidyl transferase
Proteins, 60, 2005
|
|
1NMN
 
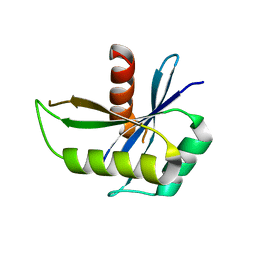 | | Structure of yqgF from Escherichia coli, a hypothetical protein | | 分子名称: | Hypothetical protein yqgF | | 著者 | Galkin, A, Sarikaya, E, Krajewski, W, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | 登録日 | 2003-01-10 | | 公開日 | 2004-03-02 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of yqgF from Escherichia coli, a hypothetical protein
To be Published
|
|
3L2E
 
 | | Glycocyamine kinase, alpha-beta heterodimer from marine worm Namalycastis sp. | | 分子名称: | Glycocyamine kinase alpha chain, Glycocyamine kinase beta chain | | 著者 | Lim, K, Pullalarevu, S, Herzberg, O. | | 登録日 | 2009-12-15 | | 公開日 | 2010-03-02 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis for the mechanism and substrate specificity of glycocyamine kinase, a phosphagen kinase family member.
Biochemistry, 49, 2010
|
|
3DBV
 
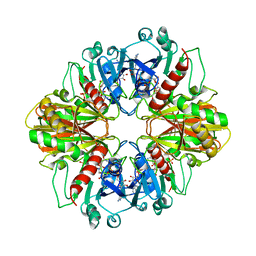 | | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE MUTANT WITH LEU 33 REPLACED BY THR, THR 34 REPLACED BY GLY, ASP 36 REPLACED BY GLY, LEU 187 REPLACED BY ALA, AND PRO 188 REPLACED BY SER COMPLEXED WITH NAD+ | | 分子名称: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | 著者 | Didierjean, C, Rahuel-Clermont, S, Vitoux, B, Dideberg, O, Branlant, G, Aubry, A. | | 登録日 | 1997-01-06 | | 公開日 | 1997-07-07 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | A crystallographic comparison between mutated glyceraldehyde-3-phosphate dehydrogenases from Bacillus stearothermophilus complexed with either NAD+ or NADP+.
J.Mol.Biol., 268, 1997
|
|
3GAY
 
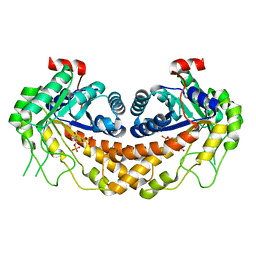 | | Structure of Giardia fructose-1,6-biphosphate aldolase in complex with tagatose-1,6-biphosphate | | 分子名称: | 1,6-di-O-phosphono-D-tagatose, Fructose-bisphosphate aldolase, ZINC ION | | 著者 | Galkin, A, Herzberg, O. | | 登録日 | 2009-02-18 | | 公開日 | 2009-03-31 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural insights into the substrate binding and stereoselectivity of giardia fructose-1,6-bisphosphate aldolase.
Biochemistry, 48, 2009
|
|
3GB6
 
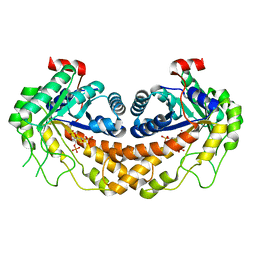 | | Structure of Giardia fructose-1,6-biphosphate aldolase D83A mutant in complex with fructose-1,6-bisphosphate | | 分子名称: | 1,6-di-O-phosphono-D-fructose, Fructose-bisphosphate aldolase, ZINC ION | | 著者 | Galkin, A, Herzberg, O. | | 登録日 | 2009-02-18 | | 公開日 | 2009-03-31 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural insights into the substrate binding and stereoselectivity of giardia fructose-1,6-bisphosphate aldolase.
Biochemistry, 48, 2009
|
|
