8BBL
 
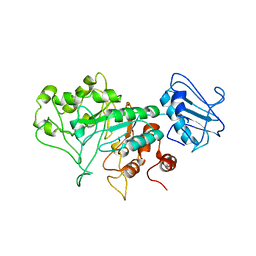 | | SGL a GH20 family sulfoglycosidase | | 分子名称: | Beta-N-acetylhexosaminidase | | 著者 | Dong, M.D, Roth, C.R, Jin, Y.J. | | 登録日 | 2022-10-13 | | 公開日 | 2023-01-11 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.711 Å) | | 主引用文献 | Mechanistic and Structural Insights into the Specificity and Biological Functions of Bacterial Sulfoglycosidases
Acs Catalysis, 2022
|
|
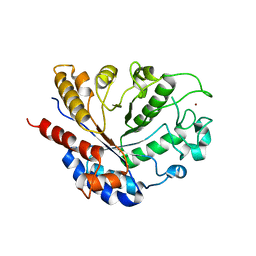
8BAL
 
 | |
5MX7
 
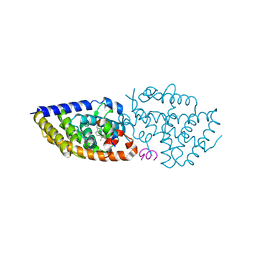 | | 1a,20S-dihydroxyvitamin D3 VDR complex | | 分子名称: | 1a,20S-dihydroxyvitamin D3, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | 著者 | Rochel, N, Belorusova, A.Y. | | 登録日 | 2017-01-21 | | 公開日 | 2017-11-29 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | 1 alpha,20S-Dihydroxyvitamin D3 Interacts with Vitamin D Receptor: Crystal Structure and Route of Chemical Synthesis.
Sci Rep, 7, 2017
|
|
6DZF
 
 | |
6L7O
 
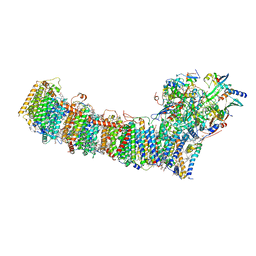 | | cryo-EM structure of cyanobacteria Fd-NDH-1L complex | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, BETA-CAROTENE, ... | | 著者 | Zhang, C, Shuai, J, Wu, J, Lei, M. | | 登録日 | 2019-11-02 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural insights into NDH-1 mediated cyclic electron transfer.
Nat Commun, 11, 2020
|
|
6L7P
 
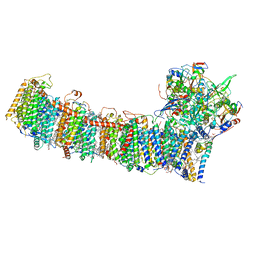 | | cryo-EM structure of cyanobacteria NDH-1LdelV complex | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, BETA-CAROTENE, ... | | 著者 | Zhang, C, Shuai, J, Wu, J, Lei, M. | | 登録日 | 2019-11-02 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural insights into NDH-1 mediated cyclic electron transfer.
Nat Commun, 11, 2020
|
|
8IOY
 
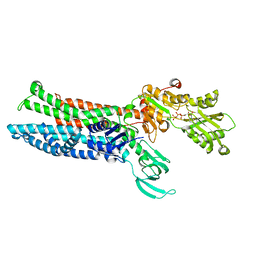 | | Structure of ATP7B C983S/C985S/D1027A mutant with AMP-PNP | | 分子名称: | Copper-transporting ATPase 2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Yang, G, Xu, L, Guo, J, Wu, Z. | | 登録日 | 2023-03-13 | | 公開日 | 2023-04-26 | | 最終更新日 | 2023-05-03 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structures of the human Wilson disease copper transporter ATP7B.
Cell Rep, 42, 2023
|
|
5BTU
 
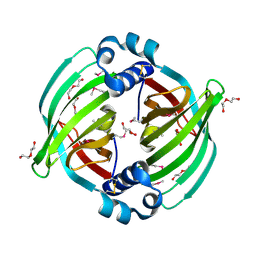 | |
5BU3
 
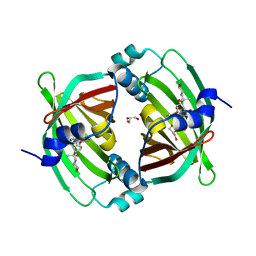 | | Crystal Structure of Diels-Alderase PyrI4 in complex with its product | | 分子名称: | (4S,4aS,6aS,8R,9R,10aR,13R,14aS,18aR,18bR)-9-ethyl-4,8,19-trihydroxy-10a,12,13,18a-tetramethyl-2,3,4,4a,5,6,6a,7,8,9,10,10a,13,14,18a,18b-hexadecahydro-1H-14a,17-(metheno)benzo[b]naphtho[2,1-h]azacyclododecine-16,18(15H,17H)-dione, GLYCEROL, PyrI4 | | 著者 | Pan, L, Guo, Y, Liu, J. | | 登録日 | 2015-06-03 | | 公開日 | 2016-02-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.897 Å) | | 主引用文献 | Enzyme-Dependent [4 + 2] Cycloaddition Depends on Lid-like Interaction of the N-Terminal Sequence with the Catalytic Core in PyrI4
Cell Chem Biol, 23, 2016
|
|
6Y7V
 
 | |
7P5E
 
 | | Pyrazole Carboxylic Acid Inhibitors of KEAP1:NRF2 interaction | | 分子名称: | 1-[6-[3-(dimethylcarbamoyl)phenyl]pyridin-2-yl]-5-(trifluoromethyl)pyrazole-4-carboxylic acid, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Davies, T.G, Cleasby, A. | | 登録日 | 2021-07-14 | | 公開日 | 2021-11-17 | | 最終更新日 | 2021-11-24 | | 実験手法 | X-RAY DIFFRACTION (1.874 Å) | | 主引用文献 | Fragment-Guided Discovery of Pyrazole Carboxylic Acid Inhibitors of the Kelch-like ECH-Associated Protein 1: Nuclear Factor Erythroid 2 Related Factor 2 (KEAP1:NRF2) Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
7P58
 
 | | Pyrazole Carboxylic Acid Inhibitors of KEAP1:NRF2 interaction | | 分子名称: | 1-[6-(3-propan-2-yloxyphenyl)pyridin-2-yl]-5-(trifluoromethyl)pyrazole-4-carboxylic acid, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Davies, T.G. | | 登録日 | 2021-07-14 | | 公開日 | 2021-11-17 | | 最終更新日 | 2021-11-24 | | 実験手法 | X-RAY DIFFRACTION (1.886 Å) | | 主引用文献 | Fragment-Guided Discovery of Pyrazole Carboxylic Acid Inhibitors of the Kelch-like ECH-Associated Protein 1: Nuclear Factor Erythroid 2 Related Factor 2 (KEAP1:NRF2) Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
7P5N
 
 | | Pyrazole Carboxylic Acid Inhibitors of KEAP1:NRF2 interaction | | 分子名称: | 1-[3-[(1~{R},3~{S})-3-[(2~{R})-2-butylpyrrolidin-1-yl]carbonylcyclohexyl]phenyl]-5-cyclopropyl-pyrazole-4-carboxylic acid, CHLORIDE ION, Kelch-like ECH-associated protein 1 | | 著者 | Davies, T.G. | | 登録日 | 2021-07-14 | | 公開日 | 2021-11-17 | | 最終更新日 | 2021-11-24 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Fragment-Guided Discovery of Pyrazole Carboxylic Acid Inhibitors of the Kelch-like ECH-Associated Protein 1: Nuclear Factor Erythroid 2 Related Factor 2 (KEAP1:NRF2) Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
7P5P
 
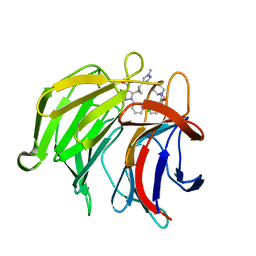 | | Pyrazole Carboxylic Acid Inhibitors of KEAP1:NRF2 interaction | | 分子名称: | 5-[(1~{R},2~{R})-2-(1-methyl-1,2,3-triazol-4-yl)cyclopropyl]-1-[3-[3-[(2~{R})-2-propylpiperidin-1-yl]carbonylphenyl]phenyl]pyrazole-4-carboxylic acid, CHLORIDE ION, Kelch-like ECH-associated protein 1, ... | | 著者 | Davies, T.G. | | 登録日 | 2021-07-14 | | 公開日 | 2021-11-17 | | 最終更新日 | 2021-11-24 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Fragment-Guided Discovery of Pyrazole Carboxylic Acid Inhibitors of the Kelch-like ECH-Associated Protein 1: Nuclear Factor Erythroid 2 Related Factor 2 (KEAP1:NRF2) Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
7P5I
 
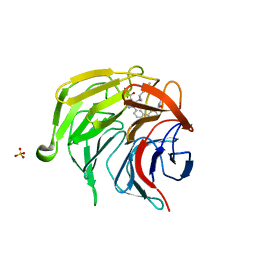 | | Pyrazole Carboxylic Acid Inhibitors of KEAP1:NRF2 interaction | | 分子名称: | 1-[3-[3-(dimethylcarbamoyl)phenyl]phenyl]-5-[(1~{R},2~{R})-2-(1-methyl-1,2,3-triazol-4-yl)cyclopropyl]pyrazole-4-carboxylic acid, CHLORIDE ION, Kelch-like ECH-associated protein 1, ... | | 著者 | Davies, T.G, Cleasby, A. | | 登録日 | 2021-07-14 | | 公開日 | 2021-11-17 | | 最終更新日 | 2021-11-24 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Fragment-Guided Discovery of Pyrazole Carboxylic Acid Inhibitors of the Kelch-like ECH-Associated Protein 1: Nuclear Factor Erythroid 2 Related Factor 2 (KEAP1:NRF2) Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
7P5F
 
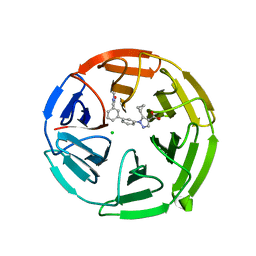 | | Pyrazole Carboxylic Acid Inhibitors of KEAP1:NRF2 interaction | | 分子名称: | 5-cyclopropyl-1-[3-[3-(dimethylcarbamoyl)phenyl]phenyl]pyrazole-4-carboxylic acid, CHLORIDE ION, Kelch-like ECH-associated protein 1 | | 著者 | Davies, T.G. | | 登録日 | 2021-07-14 | | 公開日 | 2021-11-17 | | 最終更新日 | 2021-11-24 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Fragment-Guided Discovery of Pyrazole Carboxylic Acid Inhibitors of the Kelch-like ECH-Associated Protein 1: Nuclear Factor Erythroid 2 Related Factor 2 (KEAP1:NRF2) Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
7P5K
 
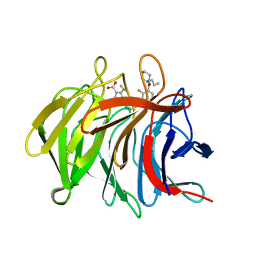 | | Pyrazole Carboxylic Acid Inhibitors of KEAP1:NRF2 interaction | | 分子名称: | 5-cyclopropyl-1-[3-[2-fluoranyl-3-[(2~{R})-2-propylpiperidin-1-yl]carbonyl-phenyl]phenyl]pyrazole-4-carboxylic acid, CHLORIDE ION, Kelch-like ECH-associated protein 1 | | 著者 | Davies, T.G. | | 登録日 | 2021-07-14 | | 公開日 | 2021-11-17 | | 最終更新日 | 2021-11-24 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Fragment-Guided Discovery of Pyrazole Carboxylic Acid Inhibitors of the Kelch-like ECH-Associated Protein 1: Nuclear Factor Erythroid 2 Related Factor 2 (KEAP1:NRF2) Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
2JR7
 
 | |
8X5Y
 
 | |
8X63
 
 | |
8X64
 
 | |
4WKN
 
 | | Crystal structure of Helicobacter pylori 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with methylthio-DADMe-Immucillin-A | | 分子名称: | (3R,4S)-1-[(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)METHYL]-4-[(METHYLSULFANYL)METHYL]PYRROLIDIN-3-OL, Aminodeoxyfutalosine nucleosidase | | 著者 | Cameron, S.A, Wang, S, Almo, S.C, Schramm, V.L. | | 登録日 | 2014-10-02 | | 公開日 | 2015-11-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | New Antibiotic Candidates against Helicobacter pylori.
J.Am.Chem.Soc., 137, 2015
|
|
4WKO
 
 | | Crystal structure of Helicobacter pylori 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with hydroxybutylthio-DADMe-Immucillin-A | | 分子名称: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-{[(4-hydroxybutyl)sulfanyl]methyl}pyrrolidin-3-ol, Aminodeoxyfutalosine nucleosidase | | 著者 | Cameron, S.A, Wang, S, Almo, S.C, Schramm, V.L. | | 登録日 | 2014-10-02 | | 公開日 | 2015-11-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | New Antibiotic Candidates against Helicobacter pylori.
J.Am.Chem.Soc., 137, 2015
|
|
4WKP
 
 | | Crystal structure of Helicobacter pylori 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with 2-(2-hydroxyethoxy)ethylthiomethyl-DADMe-Immucillin-A | | 分子名称: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-(2-{[2-(2-hydroxyethoxy)ethyl]sulfanyl}ethyl)pyrrolidin-3-ol, Aminodeoxyfutalosine nucleosidase, SULFATE ION | | 著者 | Cameron, S.A, Wang, S, Almo, S.C, Schramm, V.L. | | 登録日 | 2014-10-02 | | 公開日 | 2015-11-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | New Antibiotic Candidates against Helicobacter pylori.
J.Am.Chem.Soc., 137, 2015
|
|
5M5A
 
 | | Crystal structure of MELK in complex with an inhibitor | | 分子名称: | CHLORIDE ION, K-252A, Maternal embryonic leucine zipper kinase, ... | | 著者 | Canevari, G, Re Depaolini, S, Casale, E, Felder, E, Kuster, B, Heinzlmeir, S. | | 登録日 | 2016-10-21 | | 公開日 | 2017-12-06 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The target landscape of clinical kinase drugs.
Science, 358, 2017
|
|
