4XZA
 
 | | The crystal structure of Erve virus nucleoprotein | | 分子名称: | Nucleoprotein | | 著者 | Guo, Y, Wang, W, Liu, X, Wang, X, Wang, J, Huo, T, Liu, B. | | 登録日 | 2015-02-04 | | 公開日 | 2015-09-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural and Functional Diversity of Nairovirus-Encoded Nucleoproteins.
J.Virol., 89, 2015
|
|
4XZ8
 
 | | The crystal structure of Erve virus nucleoprotein | | 分子名称: | Nucleoprotein | | 著者 | Guo, Y, Wang, W, Liu, X, Wang, X, Wang, J, Huo, T, Liu, B. | | 登録日 | 2015-02-04 | | 公開日 | 2015-09-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structural and Functional Diversity of Nairovirus-Encoded Nucleoproteins.
J.Virol., 89, 2015
|
|
8GU4
 
 | |
8GU5
 
 | | Wild type poly(ethylene terephthalate) hydrolase | | 分子名称: | Poly(ethylene terephthalate) hydrolase | | 著者 | Xiao, Y.J, Wang, Z.F. | | 登録日 | 2022-09-09 | | 公開日 | 2022-11-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Biodegradation of highly crystallized poly(ethylene terephthalate) through cell surface codisplay of bacterial PETase and hydrophobin.
Nat Commun, 13, 2022
|
|
9BDU
 
 | | NF-kappaB RelA homo-dimer bound to AT-centric kappaB DNA | | 分子名称: | DNA (5'-D(P*AP*CP*TP*GP*GP*GP*AP*AP*AP*TP*TP*CP*CP*AP*GP*TP*GP*AP*T)-3'), DNA (5'-D(P*AP*TP*CP*AP*CP*TP*GP*GP*AP*AP*TP*TP*TP*CP*CP*CP*AP*GP*T)-3'), SULFATE ION, ... | | 著者 | Biswas, T, Huang, D, Ghosh, G. | | 登録日 | 2024-04-12 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8XS5
 
 | |
8XW2
 
 | |
8XW3
 
 | |
9BDX
 
 | | NF-kappaB RelA homo-dimer bound to CG-centric kappaB DNA | | 分子名称: | DNA (5'-D(*AP*TP*CP*AP*CP*TP*GP*GP*AP*AP*GP*TP*TP*CP*CP*CP*AP*GP*T)-3'), DNA (5'-D(P*AP*CP*TP*GP*GP*GP*AP*AP*CP*TP*TP*CP*CP*AP*GP*TP*GP*AP*T)-3'), Transcription factor p65 | | 著者 | Biswas, T, Shahabi, S, Tsodikov, O.V, Ghosh, G. | | 登録日 | 2024-04-12 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9BDV
 
 | | NF-kappaB RelA homo-dimer bound to TA-centric kappaB DNA | | 分子名称: | DNA (5'-D(*AP*TP*CP*AP*CP*TP*GP*GP*AP*AP*AP*TP*TP*CP*CP*CP*AP*GP*T)-3'), DNA (5'-D(P*AP*CP*TP*GP*GP*GP*AP*AP*TP*TP*TP*CP*CP*AP*GP*TP*GP*AP*T)-3'), SULFATE ION, ... | | 著者 | Biswas, T, Shahabi, S, Tsodikov, O.V. | | 登録日 | 2024-04-12 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9BE0
 
 | | GC-centric NF-kappaB RelA binding DNA | | 分子名称: | ACETATE ION, DNA (5'-D(*CP*GP*GP*GP*AP*AP*GP*TP*TP*CP*CP*GP*CP*GP*GP*AP*AP*CP*TP*TP*CP*CP*CP*G)-3'), ZINC ION | | 著者 | Biswas, T, Shahabi, S, Tsodikov, O.V, Ghosh, G. | | 登録日 | 2024-04-13 | | 公開日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8XAJ
 
 | |
9BDW
 
 | | NF-kappaB RelA homo-dimer bound to GC-centric kappaB DNA | | 分子名称: | DNA (5'-D(P*AP*CP*TP*GP*GP*GP*AP*AP*GP*TP*TP*CP*CP*AP*GP*TP*GP*AP*T)-3'), DNA (5'-D(P*AP*TP*CP*AP*CP*TP*GP*GP*AP*AP*CP*TP*TP*CP*CP*CP*AP*GP*T)-3'), SULFATE ION, ... | | 著者 | Biswas, T, Shahabi, S, Tsodikov, O.V, Huang, D, Ghosh, G. | | 登録日 | 2024-04-12 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9BDY
 
 | | AT-centric NF-kappaB RelA binding DNA | | 分子名称: | ACETATE ION, DNA (5'-D(*CP*GP*GP*AP*AP*TP*TP*TP*CP*CP*CP*G)-3'), DNA (5'-D(*CP*GP*GP*GP*AP*AP*AP*TP*TP*CP*CP*G)-3'), ... | | 著者 | Biswas, T, Shahabi, S, Tsodikov, O.V, Ghosh, G. | | 登録日 | 2024-04-13 | | 公開日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7CWT
 
 | |
3J9B
 
 | | Electron cryo-microscopy of an RNA polymerase | | 分子名称: | Polymerase, Polymerase basic protein 2, RNA (5'-R(*UP*UP*UP*UP*UP*A)-3'), ... | | 著者 | Chang, S.H, Sun, D.P, Liang, H.H, Wang, J, Li, J, Guo, L, Wang, X.L, Guan, C.C, Boruah, B.M, Yuan, L.M, Feng, F, Yang, M.R, Wojdyla, J, Wang, J.W, Wang, M.T, Wang, H.W, Liu, Y.F. | | 登録日 | 2014-12-16 | | 公開日 | 2015-02-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Cryo-EM Structure of Influenza Virus RNA Polymerase Complex at 4.3 angstrom Resolution.
Mol.Cell, 2015
|
|
4FFW
 
 | | Crystal Structure of Dipeptidyl Peptidase IV (DPP4, DPP-IV, CD26) in Complex with Fab + sitagliptin | | 分子名称: | (2R)-4-OXO-4-[3-(TRIFLUOROMETHYL)-5,6-DIHYDRO[1,2,4]TRIAZOLO[4,3-A]PYRAZIN-7(8H)-YL]-1-(2,4,5-TRIFLUOROPHENYL)BUTAN-2-A MINE, Dipeptidyl peptidase 4, Fab heavy chain, ... | | 著者 | Wang, Z, Sudom, A, Walker, N.P, Min, X. | | 登録日 | 2012-06-01 | | 公開日 | 2012-12-12 | | 最終更新日 | 2021-05-19 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | An Inhibitory Antibody Against DPP IV Improves Glucose Tolerance in vivo - Validation of Large Molecule Approach for DPP IV Inhibition
To be published
|
|
2MJ8
 
 | | Solution structure of CDYL2 chromodomain | | 分子名称: | Chromodomain Y-like protein 2 | | 著者 | Qin, S, Houliston, S, Arrowsmith, C.H, Edwards, A.M, Wu, H, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2013-12-28 | | 公開日 | 2014-04-23 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
6X1C
 
 | | Tubulin-RB3_SLD-TTL in complex with compound 5j | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-chlorofuro[3,2-d]pyrimidin-4-yl)-7-methoxy-3,4-dihydroquinoxalin-2(1H)-one, CALCIUM ION, ... | | 著者 | White, S.W, Yun, M. | | 登録日 | 2020-05-18 | | 公開日 | 2021-09-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | X-ray Crystallography-Guided Design, Antitumor Efficacy, and QSAR Analysis of Metabolically Stable Cyclopenta-Pyrimidinyl Dihydroquinoxalinone as a Potent Tubulin Polymerization Inhibitor.
J.Med.Chem., 64, 2021
|
|
6X1F
 
 | | Tubulin-RB3_SLD-TTL in complex with compound 5m | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 7-methoxy-4-(2-methyl-6,7-dihydro-5H-cyclopenta[d]pyrimidin-4-yl)-3,4-dihydroquinoxalin-2(1H)-one, CALCIUM ION, ... | | 著者 | White, S.W, Yun, M. | | 登録日 | 2020-05-18 | | 公開日 | 2021-09-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | X-ray Crystallography-Guided Design, Antitumor Efficacy, and QSAR Analysis of Metabolically Stable Cyclopenta-Pyrimidinyl Dihydroquinoxalinone as a Potent Tubulin Polymerization Inhibitor.
J.Med.Chem., 64, 2021
|
|
6X1E
 
 | | Tubulin-RB3_SLD-TTL in complex with compound 5l | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-chloro-6,7-dihydro-5H-cyclopenta[d]pyrimidin-4-yl)-7-methoxy-3,4-dihydroquinoxalin-2(1H)-one, CALCIUM ION, ... | | 著者 | White, S.W, Yun, M. | | 登録日 | 2020-05-18 | | 公開日 | 2021-09-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | X-ray Crystallography-Guided Design, Antitumor Efficacy, and QSAR Analysis of Metabolically Stable Cyclopenta-Pyrimidinyl Dihydroquinoxalinone as a Potent Tubulin Polymerization Inhibitor.
J.Med.Chem., 64, 2021
|
|
6NZV
 
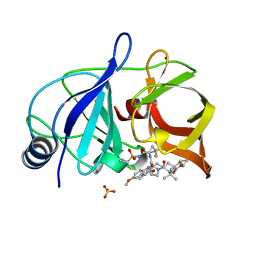 | | Crystal structure of HCV NS3/4A protease in complex with compound 12 | | 分子名称: | (1aR,5S,8S,9S,10R,22aR)-5-tert-butyl-N-[(1R,2R)-2-(difluoromethyl)-1-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}cyclopropyl]-9-ethyl-14-methoxy-3,6-dioxo-1,1a,3,4,5,6,9,10,18,19,20,21,22,22a-tetradecahydro-8H-7,10-methanocyclopropa[18,19][1,10,3,6]dioxadiazacyclononadecino[11,12-b]quinoxaline-8-carboxamide, HCV NS3/4A protease, SULFATE ION, ... | | 著者 | Appleby, T.C, Taylor, J.G. | | 登録日 | 2019-02-14 | | 公開日 | 2019-07-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Discovery of the pan-genotypic hepatitis C virus NS3/4A protease inhibitor voxilaprevir (GS-9857): A component of Vosevi®.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
8E5B
 
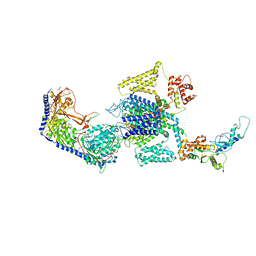 | | Human L-type voltage-gated calcium channel Cav1.3 in the presence of Amiodarone and Sofosbuvir at 3.3 Angstrom resolution | | 分子名称: | (2-butyl-1-benzofuran-3-yl){4-[2-(diethylamino)ethoxy]-3,5-diiodophenyl}methanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Gao, S, Yao, X, Yan, N. | | 登録日 | 2022-08-20 | | 公開日 | 2022-12-07 | | 最終更新日 | 2022-12-21 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis for the severe adverse interaction of sofosbuvir and amiodarone on L-type Ca v channels.
Cell, 185, 2022
|
|
8E59
 
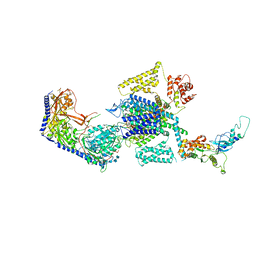 | | Human L-type voltage-gated calcium channel Cav1.3 in the presence of Amiodarone at 3.1 Angstrom resolution | | 分子名称: | (2-butyl-1-benzofuran-3-yl){4-[2-(diethylamino)ethoxy]-3,5-diiodophenyl}methanone, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Gao, S, Yao, X, Yan, N. | | 登録日 | 2022-08-20 | | 公開日 | 2022-12-07 | | 最終更新日 | 2022-12-21 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis for the severe adverse interaction of sofosbuvir and amiodarone on L-type Ca v channels.
Cell, 185, 2022
|
|
8E5A
 
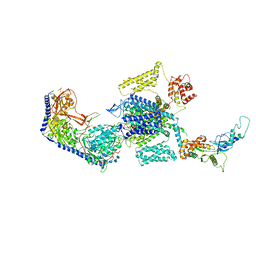 | | Human L-type voltage-gated calcium channel Cav1.3 treated with 1.4 mM Sofosbuvir at 3.3 Angstrom resolution | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Gao, S, Yao, X, Yan, N. | | 登録日 | 2022-08-20 | | 公開日 | 2022-12-07 | | 最終更新日 | 2022-12-21 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis for the severe adverse interaction of sofosbuvir and amiodarone on L-type Ca v channels.
Cell, 185, 2022
|
|
