6YBP
 
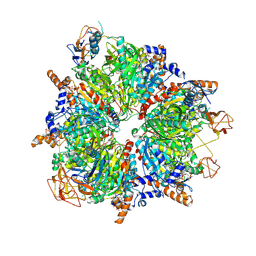 | | Propionyl-CoA carboxylase of Methylorubrum extorquens with bound CoA | | 分子名称: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, COENZYME A, Propionyl-CoA carboxylase alpha subunit, ... | | 著者 | Schuller, J.M, Schuller, S.K, Zarzycki, J, Scheffen, M, Marchal, D.M, Erb, T.J. | | 登録日 | 2020-03-17 | | 公開日 | 2020-10-28 | | 最終更新日 | 2021-05-12 | | 実験手法 | ELECTRON MICROSCOPY (3.48 Å) | | 主引用文献 | A new-to-nature carboxylation module to improve natural and synthetic CO2 fixation
Nat Catal, 2021
|
|
8VCI
 
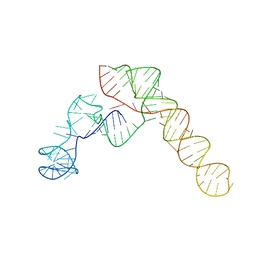 | | SARS-CoV-2 Frameshift Stimulatory Element with Upstream Multibranch Loop | | 分子名称: | Frameshift Stimulatory Element with Upstream Multi-branch Loop | | 著者 | Peterson, J.M, Becker, S.T, O'Leary, C.A, Juneja, P, Yang, Y, Moss, W.N. | | 登録日 | 2023-12-14 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (6.1 Å) | | 主引用文献 | Structure of the SARS-CoV-2 Frameshift Stimulatory Element with an Upstream Multibranch Loop.
Biochemistry, 63, 2024
|
|
6Y0C
 
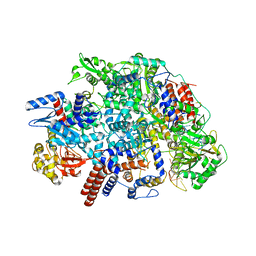 | | Influenza C virus polymerase in complex with human ANP32A - Subclass 2 | | 分子名称: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(*AP*GP*UP*AP*GP*AP*AP*AP*CP*AP*AP*GP*GP*GP*CP*CP*UP*UP*UP*U)-3'), ... | | 著者 | Fan, H, Carrique, L, Keown, J.R, Grimes, J.M, Fodor, E. | | 登録日 | 2020-02-07 | | 公開日 | 2020-11-25 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Host ANP32A mediates the assembly of the influenza virus replicase.
Nature, 587, 2020
|
|
1G6Q
 
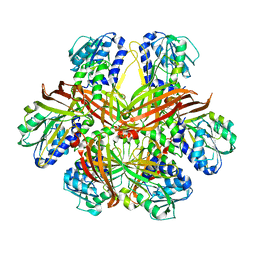 | | CRYSTAL STRUCTURE OF YEAST ARGININE METHYLTRANSFERASE, HMT1 | | 分子名称: | HNRNP ARGININE N-METHYLTRANSFERASE | | 著者 | Weiss, V.H, McBride, A.E, Soriano, M.A, Filman, D.J, Silver, P.A, Hogle, J.M. | | 登録日 | 2000-11-07 | | 公開日 | 2000-12-06 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | The structure and oligomerization of the yeast arginine methyltransferase, Hmt1.
Nat.Struct.Biol., 7, 2000
|
|
1VHR
 
 | | HUMAN VH1-RELATED DUAL-SPECIFICITY PHOSPHATASE | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HUMAN VH1-RELATED DUAL-SPECIFICITY PHOSPHATASE VHR, SULFATE ION | | 著者 | Yuvaniyama, J, Denu, J.M, Dixon, J.E, Saper, M.A. | | 登録日 | 1996-02-20 | | 公開日 | 1996-06-20 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of the dual specificity protein phosphatase VHR.
Science, 272, 1996
|
|
8V55
 
 | | Human mitochondrial DNA polymerase gamma bound to a replication fork in an open conformation | | 分子名称: | DNA polymerase subunit gamma-1, DNA polymerase subunit gamma-2, mitochondrial, ... | | 著者 | Riccio, A.A, Krahn, J.M, Bouvette, J, Borgnia, M.J, Copeland, W.C. | | 登録日 | 2023-11-30 | | 公開日 | 2024-07-10 | | 最終更新日 | 2024-07-31 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Coordinated DNA polymerization by Pol gamma and the region of LonP1 regulated proteolysis.
Nucleic Acids Res., 52, 2024
|
|
6YRS
 
 | | Structure of a new variant of GNCA ancestral beta-lactamase | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Gavira, J.A, Risso, V, Martinez-Rodriguez, S, Sanchez-Ruiz, J.M, Modi, T, Ozkan, S.B. | | 登録日 | 2020-04-20 | | 公開日 | 2021-03-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Hinge-shift mechanism as a protein design principle for the evolution of beta-lactamases from substrate promiscuity to specificity.
Nat Commun, 12, 2021
|
|
8V3N
 
 | | CCP5 in complex with Glu-P-Glu transition state analog | | 分子名称: | (2S)-2-{[(S)-[(3S)-3-acetamido-4-(ethylamino)-4-oxobutyl](hydroxy)phosphoryl]methyl}pentanedioic acid, Cytosolic carboxypeptidase-like protein 5, D-MALATE, ... | | 著者 | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | 登録日 | 2023-11-28 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V3P
 
 | | CCP5 in complex with Glu-P-peptide 2 transition state analog | | 分子名称: | Cytosolic carboxypeptidase-like protein 5, Tubulin beta-2A chain, ZINC ION | | 著者 | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | 登録日 | 2023-11-28 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V4M
 
 | | CCP5 in complex with microtubules class3 | | 分子名称: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | 登録日 | 2023-11-29 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-08-07 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V4K
 
 | | CCP5 in complex with microtubules class1 | | 分子名称: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | 登録日 | 2023-11-29 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-08-07 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V3S
 
 | | Structure of CCP5 class3 | | 分子名称: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, ZINC ION, ... | | 著者 | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | 登録日 | 2023-11-28 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-08-07 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
6YQ4
 
 | | Crystal structure of Fusobacterium nucleatum tannase | | 分子名称: | GLYCEROL, MAGNESIUM ION, SPERMIDINE, ... | | 著者 | Mancheno, J.M, Anguita, J, Rodriguez, H. | | 登録日 | 2020-04-16 | | 公開日 | 2021-03-24 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.399 Å) | | 主引用文献 | A structurally unique Fusobacterium nucleatum tannase provides detoxicant activity against gallotannins and pathogen resistance.
Microb Biotechnol, 15, 2022
|
|
8V4L
 
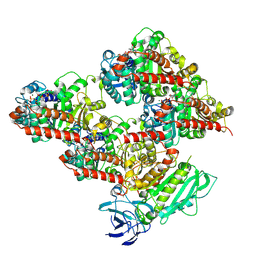 | | CCP5 in complex with microtubules class2 | | 分子名称: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | 登録日 | 2023-11-29 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-08-07 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V3Q
 
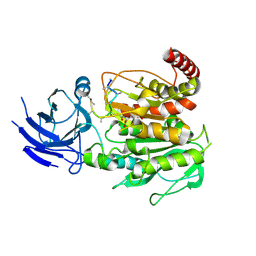 | | Structure of CCP5 class1 | | 分子名称: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, ZINC ION, ... | | 著者 | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | 登録日 | 2023-11-28 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-08-07 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V3M
 
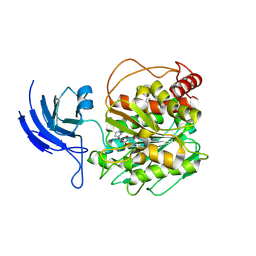 | | CCP5 apo structure | | 分子名称: | Cytosolic carboxypeptidase-like protein 5, D-MALATE, IMIDAZOLE, ... | | 著者 | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | 登録日 | 2023-11-28 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V3O
 
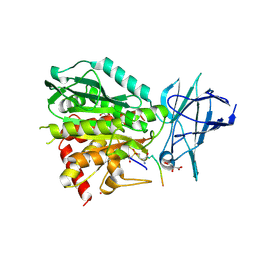 | | CCP5 in complex with Glu-P-peptide 1 transition state analog | | 分子名称: | Cytosolic carboxypeptidase-like protein 5, D-MALATE, POTASSIUM ION, ... | | 著者 | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | 登録日 | 2023-11-28 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
6YQF
 
 | |
6Y0L
 
 | | Crystal structure of human CD23 lectin domain N225D, K229E, S252N, T251N, R253G, S254G mutant | | 分子名称: | GLYCEROL, Low affinity immunoglobulin epsilon Fc receptor membrane-bound form, SULFATE ION | | 著者 | Ilkow, V.F, Davies, A.M, Sutton, B.J, McDonnell, J.M. | | 登録日 | 2020-02-09 | | 公開日 | 2021-06-16 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Reviving lost binding sites: Exploring calcium-binding site transitions between human and murine CD23.
Febs Open Bio, 11, 2021
|
|
6Y0M
 
 | | Crystal structure of human CD23 lectin domain N225D, K229E, S252N, T251N mutant | | 分子名称: | Low affinity immunoglobulin epsilon Fc receptor membrane-bound form | | 著者 | Ilkow, V.F, Davies, A.M, Sutton, B.J, McDonnell, J.M. | | 登録日 | 2020-02-09 | | 公開日 | 2021-06-16 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Reviving lost binding sites: Exploring calcium-binding site transitions between human and murine CD23.
Febs Open Bio, 11, 2021
|
|
6YD1
 
 | | SaFtsZ-DFMBA | | 分子名称: | 2,6-difluoro-3-methoxybenzamide, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | 登録日 | 2020-03-20 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Targeting the FtsZ Allosteric Binding Site with a Novel Fluorescence Polarization Screen, Cytological and Structural Approaches for Antibacterial Discovery.
J.Med.Chem., 64, 2021
|
|
6YD6
 
 | | SaFtsZ-UCM152 (comp.20) | | 分子名称: | 1,2-ETHANEDIOL, 1-methylpyrrolidin-2-one, 2,6-bis(fluoranyl)-3-[[3-(trifluoromethyl)phenyl]methoxy]benzamide, ... | | 著者 | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | 登録日 | 2020-03-20 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Targeting the FtsZ Allosteric Binding Site with a Novel Fluorescence Polarization Screen, Cytological and Structural Approaches for Antibacterial Discovery.
J.Med.Chem., 64, 2021
|
|
6YD5
 
 | | SaFtsZ-UCM151 (comp. 18) | | 分子名称: | 1,2-ETHANEDIOL, 1-methylpyrrolidin-2-one, 3-[(3-chlorophenyl)methoxy]-2,6-bis(fluoranyl)benzamide, ... | | 著者 | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | 登録日 | 2020-03-20 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Targeting the FtsZ Allosteric Binding Site with a Novel Fluorescence Polarization Screen, Cytological and Structural Approaches for Antibacterial Discovery.
J.Med.Chem., 64, 2021
|
|
2IJQ
 
 | | Crystal structure of protein rrnAC1037 from Haloarcula marismortui, Pfam DUF309 | | 分子名称: | Hypothetical protein | | 著者 | Malashkevich, V.N, Toro, R, Sauder, J.M, Schwinn, K.D, Thompson, D.A, Rutter, M.E, Dickey, M, Groshong, C, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Powell, A, Boice, A, Gheyi, T, Ozyurt, S, Atwell, S, Wasserman, S.R, Emtage, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2006-09-30 | | 公開日 | 2006-10-24 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Crystal structure of the hypothetical Protein from Haloarcula marismortui
To be Published
|
|
1XS0
 
 | | Structure of the E. coli Ivy protein | | 分子名称: | Inhibitor of vertebrate lysozyme | | 著者 | Abergel, C, Monchois, V, Byrn, D, Lazzaroni, J.C, Claverie, J.M. | | 登録日 | 2004-10-18 | | 公開日 | 2004-11-02 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Structure and evolution of the Ivy protein family, unexpected lysozyme inhibitors in Gram-negative bacteria.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
