1B0D
 
 | | Structural effects of monovalent anions on polymorphic lysozyme crystals | | 分子名称: | LYSOZYME, PARA-TOLUENE SULFONATE | | 著者 | Vaney, M.C, Broutin, I, Retailleau, P, Lafont, S, Hamiaux, C, Prange, T, Ries-Kautt, M, Ducruix, A. | | 登録日 | 1998-11-07 | | 公開日 | 1998-11-11 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Structural effects of monovalent anions on polymorphic lysozyme crystals.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1HF4
 
 | | STRUCTURAL EFFECTS OF MONOVALENT ANIONS ON POLYMORPHIC LYSOZYME CRYSTALS | | 分子名称: | LYSOZYME, NITRATE ION, SODIUM ION | | 著者 | Vaney, M.C, Broutin, I, Ries-Kautt, M, Ducruix, A. | | 登録日 | 2000-11-29 | | 公開日 | 2001-01-07 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structural Effects of Monovalent Anions on Polymorphic Lysozyme Crystals
Acta Crystallogr.,Sect.D, 57, 2001
|
|
7RB2
 
 | | Cryo-EM structure of SARS-CoV-2 NSP15 NendoU in BIS-Tris pH 6.0 | | 分子名称: | Uridylate-specific endoribonuclease | | 著者 | Godoy, A.S, Song, Y, Nakamura, A.M, Noske, G.D, Gawriljuk, V.O, Fernandes, R.S, Oliva, G. | | 登録日 | 2021-07-05 | | 公開日 | 2021-07-14 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.27 Å) | | 主引用文献 | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 51, 2023
|
|
7RB0
 
 | | Cryo-EM structure of SARS-CoV-2 NSP15 NendoU at pH 7.5 | | 分子名称: | Uridylate-specific endoribonuclease | | 著者 | Godoy, A.S, Song, Y, Nakamura, A.M, Noske, G.D, Gawriljuk, V.O, Fernandes, R.S, Oliva, G. | | 登録日 | 2021-07-05 | | 公開日 | 2021-07-14 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.98 Å) | | 主引用文献 | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 51, 2023
|
|
1LCN
 
 | | Monoclinic hen egg white lysozyme, thiocyanate complex | | 分子名称: | PROTEIN (LYSOZYME), THIOCYANATE ION | | 著者 | Hamiaux, C, Prange, T, Ducruix, A, Vaney, M.C. | | 登録日 | 1998-10-27 | | 公開日 | 1998-11-04 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Structural effects of monovalent anions on polymorphic lysozyme crystals.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
3ZR4
 
 | | STRUCTURAL EVIDENCE FOR AMMONIA TUNNELING ACROSS THE (BETA-ALPHA)8 BARREL OF THE IMIDAZOLE GLYCEROL PHOSPHATE SYNTHASE BIENZYME COMPLEX | | 分子名称: | GLUTAMINE, GLYCEROL, IMIDAZOLE GLYCEROL PHOSPHATE SYNTHASE SUBUNIT HISF, ... | | 著者 | Vega, M.C, Kuper, J, Haeger, M.C, Mohrlueder, J, Marquardt, S, Sterner, R, Wilmanns, M. | | 登録日 | 2011-06-13 | | 公開日 | 2012-10-03 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Catalysis Uncoupling in a Glutamine Amidotransferase Bienzyme by Unblocking the Glutaminase Active Site.
Chem.Biol., 19, 2012
|
|
8CIF
 
 | | Bovine naive ultralong antibody AbD08 collected at 293K | | 分子名称: | Heavy chain, Light chain | | 著者 | Clarke, J.D, Mikolajek, H, Stuart, D.I, Owens, R.J. | | 登録日 | 2023-02-09 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-07-19 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Protein-to-structure pipeline for ambient-temperature in situ crystallography at VMXi.
Iucrj, 10, 2023
|
|
7KQP
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ADP-ribose (P43 crystal form) | | 分子名称: | Non-structural protein 3, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-11-17 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (0.88 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7KEG
 
 | | Crystal structure from SARS-COV2 NendoU NSP15 | | 分子名称: | PHOSPHATE ION, Uridylate-specific endoribonuclease | | 著者 | Godoy, A.S, Nakamura, A.M, Pereira, H.M, Noske, G.D, Gawriljuk, V.O, Fernandes, R.S, Oliveira, K.I.Z, Oliva, G. | | 登録日 | 2020-10-10 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
7KEH
 
 | | Crystal structure from SARS-CoV-2 NendoU NSP15 | | 分子名称: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, Uridylate-specific endoribonuclease | | 著者 | Godoy, A.S, Nakamura, A.M, Pereira, H.M, Noske, G.D, Gawriljuk, V.O, Fernandes, R.S, Oliveira, K.I.Z, Oliva, G. | | 登録日 | 2020-10-10 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
7B6A
 
 | | BK Polyomavirus VP1 pentamer core (residues 30-299) | | 分子名称: | CALCIUM ION, DIMETHYL SULFOXIDE, IODIDE ION, ... | | 著者 | Osipov, E.M, Munawar, A, Beelen, S, Strelkov, S.V. | | 登録日 | 2020-12-07 | | 公開日 | 2022-06-22 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.44 Å) | | 主引用文献 | Discovery of novel druggable pockets on polyomavirus VP1 through crystallographic fragment-based screening to develop capsid assembly inhibitors.
Rsc Chem Biol, 3, 2022
|
|
7ME0
 
 | | Cryo-EM structure of SARS-CoV-2 NSP15 NendoU at pH 6.0 | | 分子名称: | Uridylate-specific endoribonuclease | | 著者 | Godoy, A.S, Song, Y, Nakamura, A.M, Noske, G.D, Gawriljuk, V.O, Fernandes, R.S, Oliva, G. | | 登録日 | 2021-04-06 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.48 Å) | | 主引用文献 | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
7KQW
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain (C2 crystal form, methylated) | | 分子名称: | Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-11-17 | | 公開日 | 2020-12-09 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (0.93 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7KF4
 
 | | Crystal structure from SARS-CoV-2 NendoU NSP15 | | 分子名称: | CITRIC ACID, Uridylate-specific endoribonuclease | | 著者 | Godoy, A.S, Nakamura, A.M, Pereira, H.M, Noske, G.D, Gawriljuk, V.O, Fernandes, R.S, Oliveira, K.I.Z, Oliva, G. | | 登録日 | 2020-10-13 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
7KQO
 
 | |
7KR0
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain (C2 crystal form, 100 K) | | 分子名称: | Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-11-18 | | 公開日 | 2020-12-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (0.77 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7KR1
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain (C2 crystal form, 310 K) | | 分子名称: | Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-11-18 | | 公開日 | 2020-12-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7N5Z
 
 | | SARS-CoV-2 Main protease C145S mutant | | 分子名称: | 3C-like proteinase | | 著者 | Noske, G.D, Nakamura, A.M, Gawriljuk, V.O, Lima, G.M.A, Zeri, A.C.M, Nascimento, A.F.Z, Oliva, G, Godoy, A.S. | | 登録日 | 2021-06-07 | | 公開日 | 2021-06-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | A Crystallographic Snapshot of SARS-CoV-2 Main Protease Maturation Process.
J.Mol.Biol., 433, 2021
|
|
7N6N
 
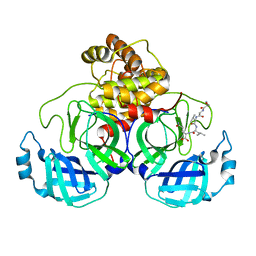 | | SARS-CoV-2 Main protease C145S mutant in complex with N and C-terminal residues | | 分子名称: | 3C-like proteinase | | 著者 | Noske, G.D, Nakamura, A.M, Gawriljuk, V.O, Lima, G.M.A, Zeri, A.C.M, Nascimento, A.F.Z, Fernandes, R.S, Oliva, G, Godoy, A.S. | | 登録日 | 2021-06-08 | | 公開日 | 2021-06-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | A Crystallographic Snapshot of SARS-CoV-2 Main Protease Maturation Process.
J.Mol.Biol., 433, 2021
|
|
7B69
 
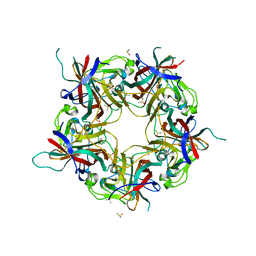 | |
7B6C
 
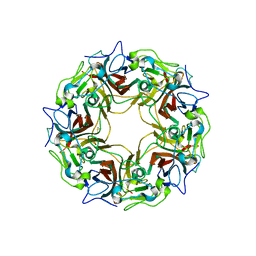 | | BK Polyomavirus VP1 pentamer fusion with long C-terminal extended arm | | 分子名称: | CALCIUM ION, DIMETHYL SULFOXIDE, Major capsid protein VP1,Major capsid protein VP1 | | 著者 | Osipov, E.M, Beelen, S, Strelkov, S.V. | | 登録日 | 2020-12-07 | | 公開日 | 2022-06-22 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.484 Å) | | 主引用文献 | Discovery of novel druggable pockets on polyomavirus VP1 through crystallographic fragment-based screening to develop capsid assembly inhibitors.
Rsc Chem Biol, 3, 2022
|
|
6G2W
 
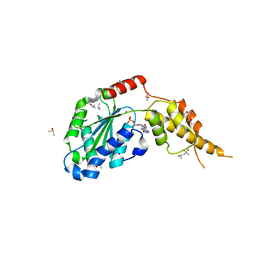 | | Crystal structure of the p97 D2 domain in a helical split-washer conformation | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, DIMETHYL SULFOXIDE, ... | | 著者 | Stach, L, Morgan, R.M.L, Freemont, P.S. | | 登録日 | 2018-03-23 | | 公開日 | 2019-04-10 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.678 Å) | | 主引用文献 | Crystal structure of the catalytic D2 domain of the AAA+ ATPase p97 reveals a putative helical split-washer-type mechanism for substrate unfolding.
Febs Lett., 594, 2020
|
|
6G30
 
 | | Crystal structure of the p97 D2 domain in a helical split-washer conformation | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, DIMETHYL SULFOXIDE, ... | | 著者 | Stach, L, Morgan, R.M.L, Freemont, P.S. | | 登録日 | 2018-03-23 | | 公開日 | 2019-04-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.418 Å) | | 主引用文献 | Crystal structure of the catalytic D2 domain of the AAA+ ATPase p97 reveals a putative helical split-washer-type mechanism for substrate unfolding.
Febs Lett., 594, 2020
|
|
6G2Z
 
 | | Crystal structure of the p97 D2 domain in a helical split-washer conformation | | 分子名称: | (3-phenyl-1,2-oxazol-5-yl)methylazanium, (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Stach, L, Morgan, R.M.L, Freemont, P.S. | | 登録日 | 2018-03-23 | | 公開日 | 2019-04-10 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.923 Å) | | 主引用文献 | Crystal structure of the catalytic D2 domain of the AAA+ ATPase p97 reveals a putative helical split-washer-type mechanism for substrate unfolding.
Febs Lett., 594, 2020
|
|
6G2V
 
 | | Crystal structure of the p97 D2 domain in a helical split-washer conformation | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, DIMETHYL SULFOXIDE, ... | | 著者 | Stach, L, Morgan, R.M.L, Freemont, P.S. | | 登録日 | 2018-03-23 | | 公開日 | 2019-04-10 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.903 Å) | | 主引用文献 | Crystal structure of the catalytic D2 domain of the AAA+ ATPase p97 reveals a putative helical split-washer-type mechanism for substrate unfolding.
Febs Lett., 594, 2020
|
|
