6WZU
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , P3221 space group | | 分子名称: | CHLORIDE ION, GLYCEROL, Non-structural protein 3, ... | | 著者 | Osipiuk, J, Tesar, C, Endres, M, Jedrzejczak, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-05-14 | | 公開日 | 2020-05-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
8F45
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a phenyl dimethyl sulfane inhibitor (cyclopropyl ketoamide warhead) | | 分子名称: | (2-methyl-2-phenylsulfanyl-propyl) ~{N}-[(2~{S})-1-[[(2~{S},3~{S})-3-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, 3C-like proteinase | | 著者 | Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | 登録日 | 2022-11-10 | | 公開日 | 2022-11-23 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
8F46
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a dimethyl phenyl sulfane inhibitor (cyano warhead) | | 分子名称: | 3C-like proteinase, N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-{[2-methyl-2-(phenylsulfanyl)propoxy]carbonyl}-L-leucinamide, TETRAETHYLENE GLYCOL | | 著者 | Liu, L, Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | 登録日 | 2022-11-10 | | 公開日 | 2022-11-23 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
2KZ3
 
 | |
2FPO
 
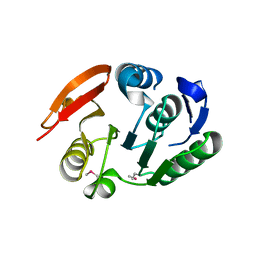 | | Putative methyltransferase yhhF from Escherichia coli. | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, methylase yhhF | | 著者 | Osipiuk, J, Kim, Y, Sanishvili, R, Skarina, T, Evdokimova, E, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2006-01-16 | | 公開日 | 2006-02-28 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Methyltransferase that modifies guanine 966 of the 16 S rRNA: functional identification and tertiary structure.
J.Biol.Chem., 282, 2007
|
|
3GN5
 
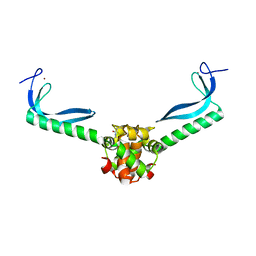 | | Structure of the E. coli protein MqsA (YgiT/b3021) | | 分子名称: | GLYCEROL, HTH-type transcriptional regulator MQSA (YGIT/b3021), ZINC ION | | 著者 | Brown, B.L, Arruda, J.M, Peti, W, Page, R. | | 登録日 | 2009-03-16 | | 公開日 | 2010-01-12 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Three dimensional structure of the MqsR:MqsA complex: a novel TA pair comprised of a toxin homologous to RelE and an antitoxin with unique properties.
Plos Pathog., 5, 2009
|
|
3HYK
 
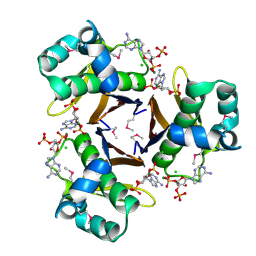 | | 2.31 Angstrom resolution crystal structure of a holo-(acyl-carrier-protein) synthase from Bacillus anthracis str. Ames in complex with CoA (3',5'-ADP) | | 分子名称: | ADENOSINE-3'-5'-DIPHOSPHATE, CHLORIDE ION, Holo-[acyl-carrier-protein] synthase, ... | | 著者 | Halavaty, A.S, Minasov, G, Skarina, T, Onopriyenko, O, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2009-06-22 | | 公開日 | 2009-06-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Structural characterization and comparison of three acyl-carrier-protein synthases from pathogenic bacteria.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3HI2
 
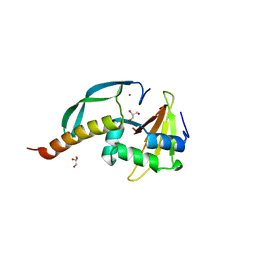 | | Structure of the N-terminal domain of the E. coli antitoxin MqsA (YgiT/b3021) in complex with the E. coli toxin MqsR (YgiU/b3022) | | 分子名称: | GLYCEROL, HTH-type transcriptional regulator mqsA(ygiT), Motility quorum-sensing regulator mqsR, ... | | 著者 | Grigoriu, S, Brown, B.L, Arruda, J.M, Peti, W, Page, R. | | 登録日 | 2009-05-18 | | 公開日 | 2010-01-12 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Three dimensional structure of the MqsR:MqsA complex: a novel TA pair comprised of a toxin homologous to RelE and an antitoxin with unique properties.
Plos Pathog., 5, 2009
|
|
1TF1
 
 | | Crystal Structure of the E. coli Glyoxylate Regulatory Protein Ligand Binding Domain | | 分子名称: | Negative regulator of allantoin and glyoxylate utilization operons | | 著者 | Walker, J.R, Skarina, T, Kudrytska, M, Joachimiak, A, Arrowsmith, C, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2004-05-26 | | 公開日 | 2004-08-03 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural and biochemical study of effector molecule recognition by the E.coli glyoxylate and allantoin utilization regulatory protein AllR.
J.Mol.Biol., 358, 2006
|
|
7TUM
 
 | | Multi-Hit SFX using MHz XFEL sources- first hit | | 分子名称: | 1,2-ETHANEDIOL, Lysozyme C, SODIUM ION | | 著者 | Darmanin, C, Holmes, S, Abbey, B. | | 登録日 | 2022-02-03 | | 公開日 | 2022-07-20 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.202 Å) | | 主引用文献 | Megahertz pulse trains enable multi-hit serial femtosecond crystallography experiments at X-ray free electron lasers.
Nat Commun, 13, 2022
|
|
7M6B
 
 | | The Crystal Structure of Mcbe1 | | 分子名称: | 1,2-ETHANEDIOL, S-ADENOSYL-L-HOMOCYSTEINE, S-ADENOSYLMETHIONINE, ... | | 著者 | Alahuhta, P.M, Lunin, V.V. | | 登録日 | 2021-03-25 | | 公開日 | 2021-04-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Target highlights in CASP14: Analysis of models by structure providers.
Proteins, 89, 2021
|
|
4ZDN
 
 | | Streptomyces platensis isomigrastatin ketosynthase domain MgsF KS4 | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AT-less polyketide synthase, CHLORIDE ION | | 著者 | Chang, C, Li, H, Endres, M, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2015-04-17 | | 公開日 | 2015-05-13 | | 最終更新日 | 2023-03-22 | | 実験手法 | X-RAY DIFFRACTION (2.509 Å) | | 主引用文献 | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5CJO
 
 | |
5BS6
 
 | | Apo structure of transcriptional factor AraR from Bacteroides thetaiotaomicron VPI | | 分子名称: | 1,2-ETHANEDIOL, transcriptional regulator AraR | | 著者 | Chang, C, Tesar, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-06-01 | | 公開日 | 2015-06-17 | | 最終更新日 | 2015-12-16 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | A novel transcriptional regulator of L-arabinose utilization in human gut bacteria.
Nucleic Acids Res., 43, 2015
|
|
6AZ2
 
 | |
6AYZ
 
 | |
5DD4
 
 | | Apo structure of transcriptional factor AraR from Bacteroides thetaiotaomicron VPI | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, TRANSCRIPTIONAL REGULATOR AraR | | 著者 | Chang, C, Tesar, C, Rodionov, D, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-08-24 | | 公開日 | 2015-09-09 | | 最終更新日 | 2015-12-16 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | A novel transcriptional regulator of L-arabinose utilization in human gut bacteria.
Nucleic Acids Res., 43, 2015
|
|
5DDG
 
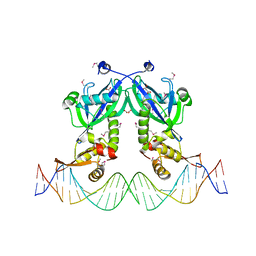 | | The structure of transcriptional factor AraR from Bacteroides thetaiotaomicron VPI in complex with target double strand DNA | | 分子名称: | DNA (27-MER), FORMIC ACID, MALONIC ACID, ... | | 著者 | Chang, C, Tesar, C, Rodionov, D, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-08-24 | | 公開日 | 2015-09-09 | | 最終更新日 | 2015-12-16 | | 実験手法 | X-RAY DIFFRACTION (3.06 Å) | | 主引用文献 | A novel transcriptional regulator of L-arabinose utilization in human gut bacteria.
Nucleic Acids Res., 43, 2015
|
|
5DEQ
 
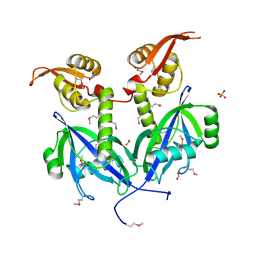 | | Crystal structure of transcriptional factor AraR from Bacteroides thetaiotaomicron VPI in complex with L-arabinose | | 分子名称: | FORMIC ACID, SULFATE ION, TRANSCRIPTIONAL REGULATOR AraR, ... | | 著者 | Chang, C, Tesar, C, Rodionov, D, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-08-25 | | 公開日 | 2015-10-21 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | A novel transcriptional regulator of L-arabinose utilization in human gut bacteria.
Nucleic Acids Res., 43, 2015
|
|
2BZ9
 
 | |
6UG1
 
 | |
2B23
 
 | | Human estrogen receptor alpha ligand-binding domain and a glucocorticoid receptor-interacting protein 1 NR box II peptide | | 分子名称: | Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Rajan, S.S, Hsieh, R.W, Sharma, S.K, Greene, G.L. | | 登録日 | 2005-09-16 | | 公開日 | 2006-09-19 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | NFkappaB selectivity of estrogen receptor ligands revealed by comparative crystallographic analyses
Nat.Chem.Biol., 4, 2008
|
|
4HRE
 
 | | Crystal Structure of p11/Annexin A2 Heterotetramer in Complex with SMARCA3 Peptide | | 分子名称: | Annexin A2, Helicase-like transcription factor, Protein S100-A10 | | 著者 | Gao, P, Patel, D.J. | | 登録日 | 2012-10-27 | | 公開日 | 2013-03-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.7852 Å) | | 主引用文献 | SMARCA3, a Chromatin-Remodeling Factor, Is Required for p11-Dependent Antidepressant Action.
Cell(Cambridge,Mass.), 152, 2013
|
|
4HRH
 
 | | Crystal Structure of p11-Annexin A2(N-terminal) Fusion Protein in Complex with SMARCA3 Peptide | | 分子名称: | Helicase-like transcription factor, Protein S100-A10, Annexin A2, ... | | 著者 | Gao, P, Patel, D.J. | | 登録日 | 2012-10-27 | | 公開日 | 2013-03-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.001 Å) | | 主引用文献 | SMARCA3, a Chromatin-Remodeling Factor, Is Required for p11-Dependent Antidepressant Action.
Cell(Cambridge,Mass.), 152, 2013
|
|
4HRG
 
 | |
