8IPT
 
 | |
8IPS
 
 | |
8IPR
 
 | |
8IPQ
 
 | |
8IV5
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 8H12 and 1C4 (local refinement) | | 分子名称: | Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, heavy chain of 1C4, ... | | 著者 | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | 登録日 | 2023-03-26 | | 公開日 | 2023-08-16 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (3.77 Å) | | 主引用文献 | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8IV8
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 3E2 and 1C4 (local refinement) | | 分子名称: | Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, heavy chain of 1C4, ... | | 著者 | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | 登録日 | 2023-03-26 | | 公開日 | 2023-08-16 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (3.92 Å) | | 主引用文献 | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8IVA
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs XMA01 and 3E2 (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, heavy chain of 3E2, ... | | 著者 | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | 登録日 | 2023-03-26 | | 公開日 | 2023-08-16 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (3.95 Å) | | 主引用文献 | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8IV4
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 8H12 and 3E2 (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, heavy chain of 3E2, ... | | 著者 | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | 登録日 | 2023-03-26 | | 公開日 | 2023-08-16 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (3.59 Å) | | 主引用文献 | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
7M0I
 
 | |
6KHR
 
 | |
7LBQ
 
 | |
7LBK
 
 | |
7LBP
 
 | |
7LBO
 
 | |
8IXI
 
 | |
3PE6
 
 | |
5F66
 
 | |
5F6M
 
 | | Isotropic Trypsin Model for Comparison of Diffuse Scattering | | 分子名称: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | 著者 | Van Benschoten, A.H, Wall, M.E, Fraser, J.S. | | 登録日 | 2015-12-06 | | 公開日 | 2016-01-06 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Measuring and modeling diffuse scattering in protein X-ray crystallography.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6JIV
 
 | | SspE crystal structure | | 分子名称: | SspE protein | | 著者 | Bing, Y.Z, Yang, H.G. | | 登録日 | 2019-02-23 | | 公開日 | 2020-03-25 | | 最終更新日 | 2020-07-08 | | 実験手法 | X-RAY DIFFRACTION (3.31 Å) | | 主引用文献 | SspABCD-SspE is a phosphorothioation-sensing bacterial defence system with broad anti-phage activities.
Nat Microbiol, 5, 2020
|
|
4M36
 
 | |
6ISU
 
 | | Crystal structure of Lys27-linked di-ubiquitin in complex with its selective interacting protein UCHL3 | | 分子名称: | Ubiquitin, Ubiquitin carboxyl-terminal hydrolase isozyme L3 | | 著者 | Ding, S, Pan, M, Zheng, Q, Ren, Y, Hong, D. | | 登録日 | 2018-11-19 | | 公開日 | 2019-02-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.866 Å) | | 主引用文献 | Chemical Protein Synthesis Enabled Mechanistic Studies on the Molecular Recognition of K27-linked Ubiquitin Chains.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
6K6L
 
 | | YGL082W-catalytic domain | | 分子名称: | pseudo deubiquitinase | | 著者 | Lu, L.N, Wang, F. | | 登録日 | 2019-06-03 | | 公開日 | 2019-07-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Inactivity of YGL082W in vitro due to impairment of conformational change in the catalytic center loop
Sci China Chem, 2019
|
|
8PVM
 
 | | formaldehyde-inhibited [FeFe]-hydrogenase CpI from Clostridium pasteurianum, variant C299D | | 分子名称: | CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, FORMYL GROUP, ... | | 著者 | Duan, J, Hofmann, E, Happe, T. | | 登録日 | 2023-07-18 | | 公開日 | 2023-11-29 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Insights into the Molecular Mechanism of Formaldehyde Inhibition of [FeFe]-Hydrogenases.
J.Am.Chem.Soc., 145, 2023
|
|
8QM3
 
 | |
4L6Q
 
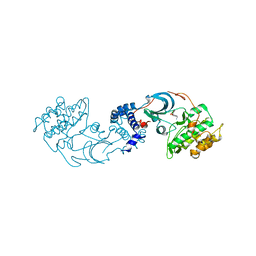 | | ROCK2 in complex with benzoxaborole | | 分子名称: | 6-[4-(aminomethyl)-2-fluorophenoxy]-2,1-benzoxaborol-1(3H)-ol, Rho-associated protein kinase 2 | | 著者 | Rock, F, Jarnagin, K. | | 登録日 | 2013-06-12 | | 公開日 | 2013-10-02 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Linking phenotype to kinase: identification of a novel benzoxaborole hinge-binding motif for kinase inhibition and development of high-potency rho kinase inhibitors.
J.Pharmacol.Exp.Ther., 347, 2013
|
|
