8G38
 
 | | Time-resolved cryo-EM study of the 70S recycling by the HflX:3rd Intermediate | | 分子名称: | 16S, 23S, 30S ribosomal protein S10, ... | | 著者 | Bhattacharjee, S, Brown, P.Z, Frank, J. | | 登録日 | 2023-02-07 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Time resolution in cryo-EM using a PDMS-based microfluidic chip assembly and its application to the study of HflX-mediated ribosome recycling.
Cell, 187, 2024
|
|
8G31
 
 | | Time-resolved cryo-EM study of the 70S recycling by the HflX:2nd Intermediate | | 分子名称: | 16S, 23S, 30S ribosomal protein S10, ... | | 著者 | Bhattacharjee, S, Brown, P.Z, Frank, J. | | 登録日 | 2023-02-06 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Time resolution in cryo-EM using a PDMS-based microfluidic chip assembly and its application to the study of HflX-mediated ribosome recycling.
Cell, 187, 2024
|
|
8G2U
 
 | | Time-resolved cryo-EM study of the 70S recycling by the HflX:control-apo-70S at 900ms | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Bhattacharjee, S, Brown, P.Z, Frank, J. | | 登録日 | 2023-02-06 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Time resolution in cryo-EM using a PDMS-based microfluidic chip assembly and its application to the study of HflX-mediated ribosome recycling.
Cell, 187, 2024
|
|
8G34
 
 | | Time-resolved cryo-EM study of the 70S recycling by the HflX:1st intermediate | | 分子名称: | 16S, 23S, 30S ribosomal protein S10, ... | | 著者 | Bhattacharjee, S, Brown, P.Z, Frank, J. | | 登録日 | 2023-02-06 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Time resolution in cryo-EM using a PDMS-based microfluidic chip assembly and its application to the study of HflX-mediated ribosome recycling.
Cell, 187, 2024
|
|
5USF
 
 | |
5GT5
 
 | |
8HRX
 
 | | Cryo-EM structure of human NTCP-myr-preS1-YN9048Fab complex | | 分子名称: | Fab heavy chain from antibody IgG clone number YN9048, Fab light chain from antibody IgG clone number YN9048, PreS1 protein (Fragment), ... | | 著者 | Asami, J, Shimizu, T, Ohto, U. | | 登録日 | 2022-12-16 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (2.89 Å) | | 主引用文献 | Structural basis of hepatitis B virus receptor binding.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HPA
 
 | | Monkeypox virus DNA replication holoenzyme F8, A22 and E4 complex in a DNA binding form | | 分子名称: | DNA (5'-D(*CP*GP*AP*TP*CP*CP*TP*TP*CP*CP*CP*CP*TP*AP*C)-3'), DNA (5'-D(P*AP*TP*GP*GP*TP*AP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*CP*G)-3'), DNA polymerase, ... | | 著者 | Xu, Y, Wu, Y, Zhang, Y, Fan, R, Yang, Y, Li, D, Yang, B, Zhang, Z, Dong, C. | | 登録日 | 2022-12-12 | | 公開日 | 2024-01-31 | | 実験手法 | ELECTRON MICROSCOPY (3.01 Å) | | 主引用文献 | Structure of DNA replication machinery from human monkeypox virus
To Be Published
|
|
8HRY
 
 | | Cryo-EM structure of human NTCP-myr-preS1-YN9016Fab complex | | 分子名称: | Fab heavy chain from antibody IgG clone number YN9016, Fab light chain from antibody IgG clone number YN9016, Large S protein (Fragment), ... | | 著者 | Asami, J, Shimizu, T, Ohto, U. | | 登録日 | 2022-12-16 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (3.11 Å) | | 主引用文献 | Structural basis of hepatitis B virus receptor binding.
Nat.Struct.Mol.Biol., 31, 2024
|
|
3SE9
 
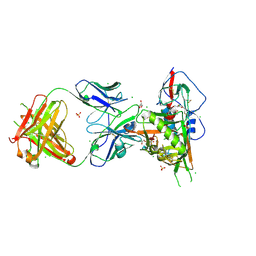 | | Crystal structure of broadly and potently neutralizing antibody VRC-PG04 in complex with HIV-1 gp120 | | 分子名称: | (R,R)-2,3-BUTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Kwong, P.D, Zhou, T. | | 登録日 | 2011-06-10 | | 公開日 | 2011-08-10 | | 最終更新日 | 2021-04-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing.
Science, 333, 2011
|
|
5DG2
 
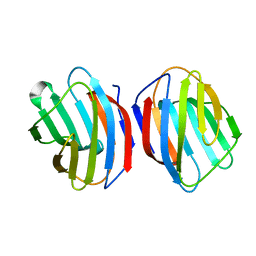 | | Sugar binding protein - human galectin-2 (dimer) | | 分子名称: | Galectin-2, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Su, J.Y, Si, Y.L. | | 登録日 | 2015-08-27 | | 公開日 | 2016-09-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.612 Å) | | 主引用文献 | Human galectin-2 interacts with carbohydrates and peptides non-classically: new insight from X-ray crystallography and hemagglutination.
Acta Biochim. Biophys. Sin. (Shanghai), 48, 2016
|
|
5DG1
 
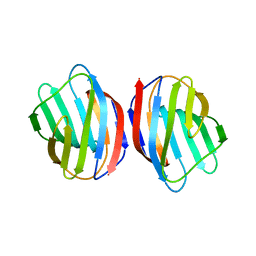 | | Sugar binding protein - human galectin-2 | | 分子名称: | Galectin-2, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Su, J.Y, Si, Y.L. | | 登録日 | 2015-08-27 | | 公開日 | 2016-09-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Human galectin-2 interacts with carbohydrates and peptides non-classically: new insight from X-ray crystallography and hemagglutination.
Acta Biochim.Biophys.Sin., 2016
|
|
4JAM
 
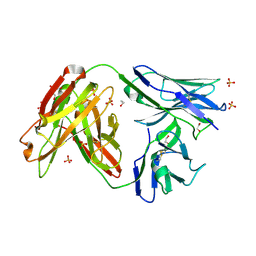 | | Crystal structure of broadly neutralizing anti-hiv-1 antibody ch103 | | 分子名称: | 1,2-ETHANEDIOL, ANTIGEN BINDING FRAGMENT OF HEAVY CHAIN of CH103, ANTIGEN BINDING FRAGMENT OF LIGHT CHAIN of CH103, ... | | 著者 | Zhou, T, Moquin, S, Zheng, A, Srivatsan, S, Kwong, P.D. | | 登録日 | 2013-02-18 | | 公開日 | 2013-04-03 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus.
Nature, 496, 2013
|
|
5VQM
 
 | |
2L89
 
 | |
5VMT
 
 | |
3DAM
 
 | | Crystal Structure of Allene oxide synthase | | 分子名称: | Cytochrome P450 74A2, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Li, L, Wang, X. | | 登録日 | 2008-05-29 | | 公開日 | 2008-09-23 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Modes of heme binding and substrate access for cytochrome P450 CYP74A revealed by crystal structures of allene oxide synthase.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3IKW
 
 | | Structure of Heparinase I from Bacteroides thetaiotaomicron | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, Heparin lyase I | | 著者 | Garron, M.L, Cygler, M, Shaya, D. | | 登録日 | 2009-08-06 | | 公開日 | 2009-09-29 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Structural snapshots of heparin depolymerization by heparin lyase I.
J.Biol.Chem., 284, 2009
|
|
3IMN
 
 | |
3ILR
 
 | | Structure of Heparinase I from Bacteroides thetaiotaomicron in complex with tetrasaccharide product | | 分子名称: | 1,2-ETHANEDIOL, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(4-1)-4-deoxy-2-O-sulfo-beta-D-erythro-hex-4-enopyranuronic acid, 4-deoxy-2-O-sulfo-beta-D-erythro-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, ... | | 著者 | Garron, M.L, Cygler, M, Shaya, D. | | 登録日 | 2009-08-07 | | 公開日 | 2009-09-29 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural snapshots of heparin depolymerization by heparin lyase I.
J.Biol.Chem., 284, 2009
|
|
3INA
 
 | | Crystal structure of heparin lyase I H151A mutant complexed with a dodecasaccharide heparin | | 分子名称: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-3,6-di-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, CALCIUM ION, Heparin lyase I | | 著者 | Han, Y.H, Ryu, K.S, Kim, H.Y, Jeon, Y.H. | | 登録日 | 2009-08-12 | | 公開日 | 2009-09-29 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural snapshots of heparin depolymerization by heparin lyase I
J.Biol.Chem., 284, 2009
|
|
8IV5
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 8H12 and 1C4 (local refinement) | | 分子名称: | Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, heavy chain of 1C4, ... | | 著者 | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | 登録日 | 2023-03-26 | | 公開日 | 2023-08-16 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (3.77 Å) | | 主引用文献 | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8IVA
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs XMA01 and 3E2 (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, heavy chain of 3E2, ... | | 著者 | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | 登録日 | 2023-03-26 | | 公開日 | 2023-08-16 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (3.95 Å) | | 主引用文献 | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8IV4
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 8H12 and 3E2 (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, heavy chain of 3E2, ... | | 著者 | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | 登録日 | 2023-03-26 | | 公開日 | 2023-08-16 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (3.59 Å) | | 主引用文献 | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8IV8
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 3E2 and 1C4 (local refinement) | | 分子名称: | Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, heavy chain of 1C4, ... | | 著者 | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | 登録日 | 2023-03-26 | | 公開日 | 2023-08-16 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (3.92 Å) | | 主引用文献 | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
