7NX6
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-222 and EY6A Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COVOX-222 Fab Heavy chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D. | | 登録日 | 2021-03-17 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Antibody evasion by the P.1 strain of SARS-CoV-2.
Cell, 184, 2021
|
|
7NX7
 
 | | Crystal structure of the K417N mutant receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-222 and EY6A Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, CITRIC ACID, ... | | 著者 | Zhou, D, Ren, J, Stuart, D. | | 登録日 | 2021-03-17 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Antibody evasion by the P.1 strain of SARS-CoV-2.
Cell, 184, 2021
|
|
7NX8
 
 | | Crystal structure of the K417T mutant receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-222 and EY6A Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, CITRIC ACID, ... | | 著者 | Zhou, D, Ren, J, Stuart, D. | | 登録日 | 2021-03-17 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Antibody evasion by the P.1 strain of SARS-CoV-2.
Cell, 184, 2021
|
|
6Z3P
 
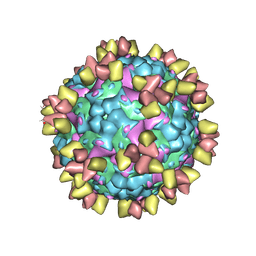 | | Structure of EV71 in complex with a protective antibody 38-3-11A Fab | | 分子名称: | SPHINGOSINE, VP1, VP2, ... | | 著者 | Zhou, D, Fry, E.E, Ren, J, Stuart, D.I. | | 登録日 | 2020-05-21 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-10-28 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural and functional analysis of protective antibodies targeting the threefold plateau of enterovirus 71.
Nat Commun, 11, 2020
|
|
6Z3Q
 
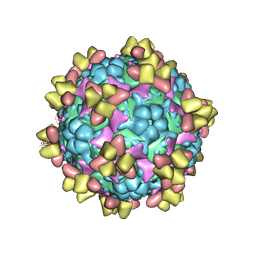 | | Structure of EV71 in complex with a protective antibody 38-1-10A Fab | | 分子名称: | Heavy chain, Light chain, SPHINGOSINE, ... | | 著者 | Zhou, D, Fry, E.E, Ren, J, Stuart, D.I. | | 登録日 | 2020-05-21 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-10-28 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Structural and functional analysis of protective antibodies targeting the threefold plateau of enterovirus 71.
Nat Commun, 11, 2020
|
|
6ZFO
 
 | | Association of two complexes of largely structurally disordered Spike ectodomain with bound EY6A Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, EY6A heavy chain, EY6A light chain, ... | | 著者 | Duyvesteyn, H.M.E, Zhou, D, Zhao, Y, Fry, E.E, Ren, J, Stuart, D.I. | | 登録日 | 2020-06-17 | | 公開日 | 2020-07-08 | | 最終更新日 | 2021-12-22 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6Z3K
 
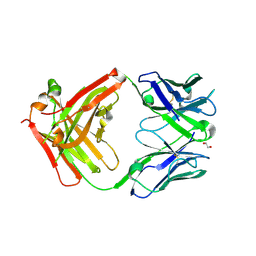 | | Structure of protective antibody 38-1-10A Fab | | 分子名称: | 1,2-ETHANEDIOL, Heavy chain, Light Chain | | 著者 | Zhou, D, Fry, E.E, Ren, J, Stuart, D.I. | | 登録日 | 2020-05-20 | | 公開日 | 2020-09-02 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural and functional analysis of protective antibodies targeting the threefold plateau of enterovirus 71.
Nat Commun, 11, 2020
|
|
8B9F
 
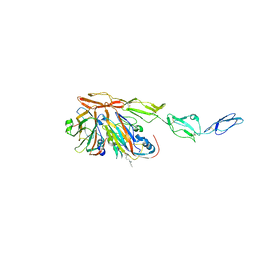 | | Structure of Echovirus 11 complexed with DAF (CD55) calculated from symmetry expansion | | 分子名称: | Complement decay-accelerating factor, Genome polyprotein, SPHINGOSINE | | 著者 | Stuart, D.I, Ren, J, Qin, L, Zhou, D. | | 登録日 | 2022-10-05 | | 公開日 | 2022-12-07 | | 最終更新日 | 2023-01-04 | | 実験手法 | ELECTRON MICROSCOPY (3.93 Å) | | 主引用文献 | Switching of Receptor Binding Poses between Closely Related Enteroviruses.
Viruses, 14, 2022
|
|
6ZDH
 
 | | SARS-CoV-2 Spike glycoprotein in complex with a neutralizing antibody EY6A Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, EY6A heavy chain, ... | | 著者 | Duyvesteyn, H.M.E, Zhou, D, Zhao, Y, Fry, E.E, Ren, J, Stuart, D.I. | | 登録日 | 2020-06-14 | | 公開日 | 2020-07-01 | | 最終更新日 | 2021-12-22 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8B8R
 
 | | Complex of Echovirus 11 with its attaching receptor decay-accelerating factor (CD55) | | 分子名称: | DECAY ACCELERATING FACTOR (CD55), SPHINGOSINE, VP1, ... | | 著者 | Stuart, D.I, Ren, J, Zhou, D, Qin, L. | | 登録日 | 2022-10-04 | | 公開日 | 2022-12-07 | | 最終更新日 | 2023-01-04 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Switching of Receptor Binding Poses between Closely Related Enteroviruses.
Viruses, 14, 2022
|
|
6ZDG
 
 | | Association of three complexes of largely structurally disordered Spike ectodomain with bound EY6A Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, EY6A heavy chain, EY6A light chain, ... | | 著者 | Duyvesteyn, H.M.E, Zhou, D, Zhao, Y, Fry, E.E, Ren, J, Stuart, D.I. | | 登録日 | 2020-06-14 | | 公開日 | 2020-07-29 | | 最終更新日 | 2021-12-22 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4ZOT
 
 | | Crystal structure of BbKI, a disulfide-free plasma kallikrein inhibitor at 1.4 A resolution | | 分子名称: | Kunitz-type serine protease inhibitor BbKI | | 著者 | Shabalin, I.G, Zhou, D, Wlodawer, A, Oliva, M.L.V. | | 登録日 | 2015-05-06 | | 公開日 | 2015-05-20 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structure of BbKI, a disulfide-free plasma kallikrein inhibitor.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
5Z3G
 
 | | Cryo-EM structure of a nucleolar pre-60S ribosome (Rpf1-TAP) | | 分子名称: | 25S rRNA, 5.8S rRNA, 60S ribosomal protein L13-A, ... | | 著者 | Zhu, X, Zhou, D, Ye, K. | | 登録日 | 2018-01-06 | | 公開日 | 2018-04-11 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.65 Å) | | 主引用文献 | Cryo-EM structure of an early precursor of large ribosomal subunit reveals a half-assembled intermediate.
Protein Cell, 10, 2019
|
|
7NEG
 
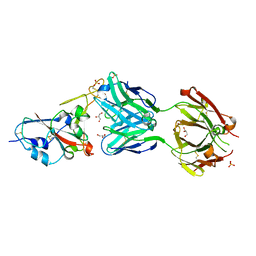 | | Crystal structure of the N501Y mutant receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-269 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody COVOX-269 Fab heavy chain, Antibody COVOX-269 Fab light chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D. | | 登録日 | 2021-02-04 | | 公開日 | 2021-03-03 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Reduced neutralization of SARS-CoV-2 B.1.1.7 variant by convalescent and vaccine sera.
Cell, 184, 2021
|
|
7NEH
 
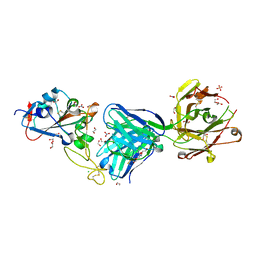 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-269 Fab | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | 著者 | Zhou, D, Ren, J, Stuart, D. | | 登録日 | 2021-02-04 | | 公開日 | 2021-03-03 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Reduced neutralization of SARS-CoV-2 B.1.1.7 variant by convalescent and vaccine sera.
Cell, 184, 2021
|
|
8R80
 
 | | SARS-CoV-2 Delta RBD in complex with XBB-9 Fab and an anti-Fab nanobody | | 分子名称: | Spike protein S1, XBB-9 Fab heavy chain, XBB-9 Fab light chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2023-11-27 | | 公開日 | 2024-05-08 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (4.03 Å) | | 主引用文献 | A structure-function analysis shows SARS-CoV-2 BA.2.86 balances antibody escape and ACE2 affinity.
Cell Rep Med, 5, 2024
|
|
6B1B
 
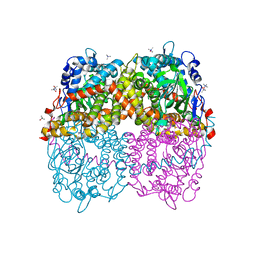 | | STRUCTURE OF 4-HYDROXYPHENYLACETATE 3-MONOOXYGENASE (HPAB), OXYGENASE COMPONENT FROM ESCHERICHIA COLI MUTANT XS6 (APO Enzyme) | | 分子名称: | 4-hydroxyphenylacetate 3-monooxygenase, oxygenase subunit, trimethylamine oxide | | 著者 | Zhou, D, Kandavelu, P, Wang, B.C, Yan, Y, Rose, J.P. | | 登録日 | 2017-09-18 | | 公開日 | 2019-05-22 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.944 Å) | | 主引用文献 | Structural Insights into Catalytic Versatility of the Flavin-dependent Hydroxylase (HpaB) from Escherichia coli.
Sci Rep, 9, 2019
|
|
4II0
 
 | |
4IHZ
 
 | |
8QZR
 
 | | SARS-CoV-2 delta RBD complexed with BA.4/5-9 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BA.4/5-9 heavy chain, BA.4/5-9 light chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2023-10-29 | | 公開日 | 2024-04-03 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.77 Å) | | 主引用文献 | Emerging variants develop total escape from potent monoclonal antibodies induced by BA.4/5 infection.
Nat Commun, 15, 2024
|
|
8QRG
 
 | | SARS-CoV-2 delta RBD complexed with XBB-2 Fab and NbC1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, NbC1, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2023-10-07 | | 公開日 | 2024-05-08 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | A structure-function analysis shows SARS-CoV-2 BA.2.86 balances antibody escape and ACE2 affinity.
Cell Rep Med, 5, 2024
|
|
8QRF
 
 | | SARS-CoV-2 delta RBD complexed with XBB-6 and beta-49 Fabs | | 分子名称: | Beta-49 heavy chain, Beta-49 light chain, Spike protein S1, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2023-10-06 | | 公開日 | 2024-05-08 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | A structure-function analysis shows SARS-CoV-2 BA.2.86 balances antibody escape and ACE2 affinity.
Cell Rep Med, 5, 2024
|
|
5C14
 
 | | Crystal structure of PECAM-1 D1D2 domain | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | 著者 | Zhou, D, Paddock, C, Newman, P, Zhu, J. | | 登録日 | 2015-06-12 | | 公開日 | 2016-01-06 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis for PECAM-1 homophilic binding.
Blood, 127, 2016
|
|
8CBE
 
 | | SARS-CoV-2 Delta-RBD complexed with BA.4/5-2 and Beta-49 Fabs | | 分子名称: | BA.4/5-2 heavy chain, BA.4/5-2 light chain, Beta-49 heavy chain, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2023-01-25 | | 公開日 | 2024-02-07 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.16 Å) | | 主引用文献 | Emerging variants develop total escape from potent monoclonal antibodies induced by BA.4/5 infection.
Nat Commun, 15, 2024
|
|
8CBF
 
 | | SARS-CoV-2 Delta-RBD complexed with Omi-42 and Beta-49 Fabs | | 分子名称: | Beta-49 heavy chain, Beta-49 light chain, CHLORIDE ION, ... | | 著者 | Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2023-01-25 | | 公開日 | 2024-02-07 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | Emerging variants develop total escape from potent monoclonal antibodies induced by BA.4/5 infection.
Nat Commun, 15, 2024
|
|
