8FE5
 
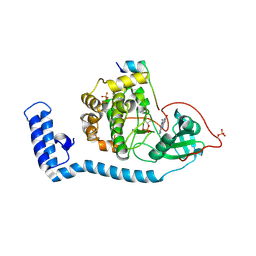 | | Structure of J-PKAc chimera complexed with Aplithianine B | | 分子名称: | 6-[(6P)-6-(1-methyl-1H-imidazol-5-yl)-2,3-dihydro-4H-1,4-thiazin-4-yl]-7,9-dihydro-8H-purin-8-one, DnaJ homolog subfamily B member 1,cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | 著者 | Du, L, Wilson, B.A.P, Li, N, Martinez Fiesco, J.A, Dalilian, M, Wang, D, Smith, E.A, Wamiru, A, Goncharova, E.I, Zhang, P, O'Keefe, B.R. | | 登録日 | 2022-12-05 | | 公開日 | 2023-10-18 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Discovery and Synthesis of a Naturally Derived Protein Kinase Inhibitor that Selectively Inhibits Distinct Classes of Serine/Threonine Kinases.
J.Nat.Prod., 86, 2023
|
|
8FAC
 
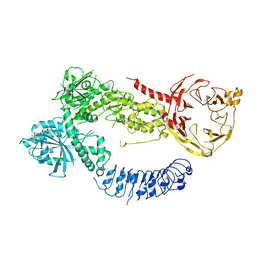 | |
8FEC
 
 | | Structure of J-PKAc chimera complexed with Aplithianine derivative | | 分子名称: | 6-[(6P)-6-(4-bromo-1-methyl-1H-imidazol-5-yl)-2,3-dihydro-4H-1,4-thiazin-4-yl]-7H-purine, DnaJ homolog subfamily B member 1,cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | 著者 | Du, L, Wilson, B.A.P, Li, N, Martinez Fiesco, J.A, Dalilian, M, Wang, D, Smith, E.A, Wamiru, A, Goncharova, E.I, Zhang, P, O'Keefe, B.R. | | 登録日 | 2022-12-06 | | 公開日 | 2023-10-18 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Discovery and Synthesis of a Naturally Derived Protein Kinase Inhibitor that Selectively Inhibits Distinct Classes of Serine/Threonine Kinases.
J.Nat.Prod., 86, 2023
|
|
8FE2
 
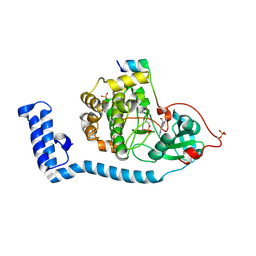 | | Structure of J-PKAc chimera complexed with Aplithianine A | | 分子名称: | 6-[(6M)-6-(1-methyl-1H-imidazol-5-yl)-2,3-dihydro-4H-1,4-thiazin-4-yl]-9H-purine, DnaJ homolog subfamily B member 1, cAMP-dependent protein kinase catalytic subunit alpha, ... | | 著者 | Du, L, Wilson, B.A.P, Li, N, Dalilian, M, Wang, D, Martinez Fiesco, J.A, Smith, E.A, Wamiru, A, Goncharova, E.I, Zhang, P, O'Keefe, B.R. | | 登録日 | 2022-12-05 | | 公開日 | 2023-10-18 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Discovery and Synthesis of a Naturally Derived Protein Kinase Inhibitor that Selectively Inhibits Distinct Classes of Serine/Threonine Kinases.
J.Nat.Prod., 86, 2023
|
|
3H4M
 
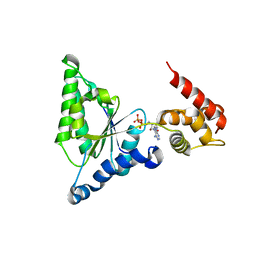 | | AAA ATPase domain of the proteasome- activating nucleotidase | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Proteasome-activating nucleotidase | | 著者 | Jeffrey, P, Zhang, F, Hu, M, Tian, G, Zhang, P, Finley, D, Shi, Y. | | 登録日 | 2009-04-20 | | 公開日 | 2009-06-09 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.106 Å) | | 主引用文献 | Structural Insights into the Regulatory Particle of the Proteasome from Methanocaldococcus jannaschii.
Mol.Cell, 34, 2009
|
|
6XF8
 
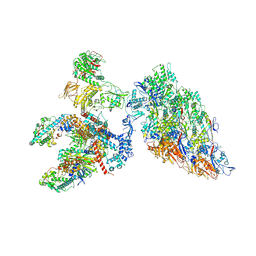 | | DLP 5 fold | | 分子名称: | Inner capsid protein lambda-1, Inner capsid protein sigma-2, Outer capsid protein mu-1, ... | | 著者 | Sutton, G, Sun, D.P, Fu, X.F, Kotecha, A, Hecksel, G.W, Clare, D.K, Zhang, P, Stuart, D, Boyce, M. | | 登録日 | 2020-06-15 | | 公開日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (6.5 Å) | | 主引用文献 | Assembly intermediates of orthoreovirus captured in the cell.
Nat Commun, 11, 2020
|
|
6Q0X
 
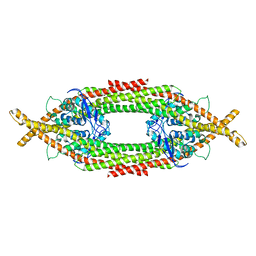 | |
6KZ8
 
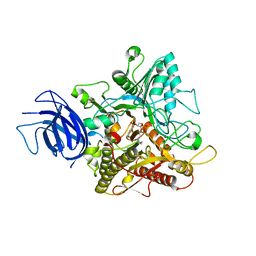 | | Crystal structure of plant Phospholipase D alpha complex with phosphatidic acid | | 分子名称: | 1,2-DIOCTANOYL-SN-GLYCERO-3-PHOSPHATE, CALCIUM ION, Phospholipase D alpha 1 | | 著者 | Li, J.X, Yu, F, Zhang, P. | | 登録日 | 2019-09-23 | | 公開日 | 2019-11-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.291 Å) | | 主引用文献 | Crystal structure of plant PLD alpha 1 reveals catalytic and regulatory mechanisms of eukaryotic phospholipase D.
Cell Res., 30, 2020
|
|
6KZ9
 
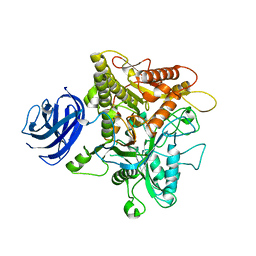 | |
8C9M
 
 | | HERV-K Gag immature lattice | | 分子名称: | Gag protein | | 著者 | Krebs, A.-S, Liu, H.-F, Zhou, Y, Rey, J.S, Levintov, L, Perilla, J.R, Bartesaghi, A, Zhang, P. | | 登録日 | 2023-01-23 | | 公開日 | 2023-02-01 | | 最終更新日 | 2023-07-05 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Molecular architecture and conservation of an immature human endogenous retrovirus.
Biorxiv, 2023
|
|
6LZ7
 
 | |
6LZ3
 
 | |
8C5V
 
 | |
2M6I
 
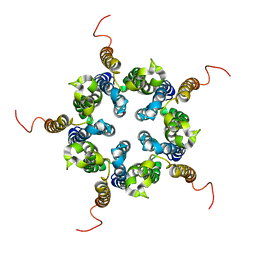 | | Putative pentameric open-channel structure of full-length transmembrane domains of human glycine receptor alpha1 subunit | | 分子名称: | Full-Length Transmembrane Domains of Human Glycine Receptor alpha1 Subunit | | 著者 | Mowrey, D, Cui, T, Jia, Y, Ma, D, Makhov, A.M, Zhang, P, Tang, P, Xu, Y. | | 登録日 | 2013-03-29 | | 公開日 | 2013-09-04 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Open-Channel Structures of the Human Glycine Receptor alpha 1 Full-Length Transmembrane Domain.
Structure, 21, 2013
|
|
2M6B
 
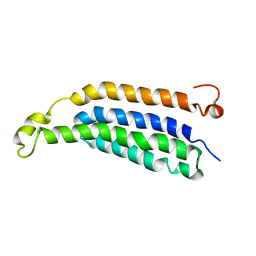 | | Structure of full-length transmembrane domains of human glycine receptor alpha1 monomer subunit | | 分子名称: | Full-Length Transmembrane Domains of Human Glycine Receptor alpha1 Subunit | | 著者 | Mowrey, D, Cui, T, Jia, Y, Ma, D, Makhov, A.M, Zhang, P, Tang, P, Xu, Y. | | 登録日 | 2013-03-28 | | 公開日 | 2013-09-04 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Open-Channel Structures of the Human Glycine Receptor alpha 1 Full-Length Transmembrane Domain.
Structure, 21, 2013
|
|
4XHR
 
 | | Structure of a phospholipid trafficking complex, native | | 分子名称: | Mitochondrial distribution and morphology protein 35, Protein UPS1, mitochondrial | | 著者 | Yu, F, He, F, Wang, C, Zhang, P. | | 登録日 | 2015-01-06 | | 公開日 | 2015-07-01 | | 最終更新日 | 2015-08-05 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structural basis of intramitochondrial phosphatidic acid transport mediated by Ups1-Mdm35 complex
Embo Rep., 16, 2015
|
|
3IWP
 
 | |
4XIZ
 
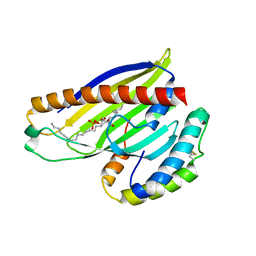 | | Structure of a phospholipid trafficking complex with substrate | | 分子名称: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, Mitochondrial distribution and morphology protein 35, Protein UPS1, ... | | 著者 | Yu, F, He, F, Wang, C, Zhang, P. | | 登録日 | 2015-01-08 | | 公開日 | 2015-07-01 | | 最終更新日 | 2015-09-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis of intramitochondrial phosphatidic acid transport mediated by Ups1-Mdm35 complex
Embo Rep., 16, 2015
|
|
6SKM
 
 | | Structure of the native full-length HIV-1 capsid protein A92E in helical assembly (-13,12) | | 分子名称: | Gag protein | | 著者 | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | 登録日 | 2019-08-16 | | 公開日 | 2020-08-26 | | 最終更新日 | 2022-03-30 | | 実験手法 | ELECTRON MICROSCOPY (4.9 Å) | | 主引用文献 | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6SLQ
 
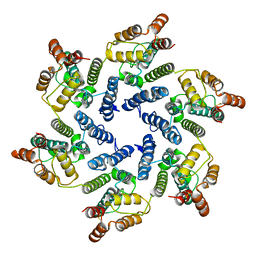 | | Structure of the native full-length HIV-1 capsid protein A92E in helical assembly (-12,11) | | 分子名称: | Gag protein | | 著者 | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | 登録日 | 2019-08-20 | | 公開日 | 2020-09-09 | | 最終更新日 | 2022-03-30 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6SLU
 
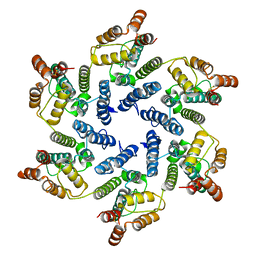 | | Structure of the native full-length HIV-1 capsid protein A92E in helical assembly (-13,11) | | 分子名称: | Gag protein | | 著者 | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | 登録日 | 2019-08-20 | | 公開日 | 2020-09-09 | | 最終更新日 | 2022-03-30 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6SMU
 
 | | Structure of the native full-length HIV-1 capsid protein in helical assembly (-13,12) | | 分子名称: | Gag protein | | 著者 | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | 登録日 | 2019-08-22 | | 公開日 | 2020-09-09 | | 最終更新日 | 2022-03-30 | | 実験手法 | ELECTRON MICROSCOPY (5 Å) | | 主引用文献 | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6SKN
 
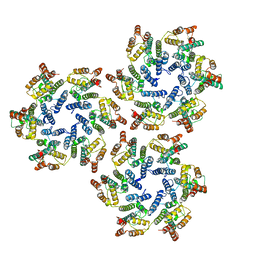 | | Structure of the native full-length HIV-1 capsid protein in helical assembly (-13,8) | | 分子名称: | Gag protein | | 著者 | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | 登録日 | 2019-08-16 | | 公開日 | 2020-08-26 | | 最終更新日 | 2022-03-30 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
3J3Y
 
 | |
6SKK
 
 | | Structure of the native full-length HIV-1 capsid protein in helical assembly (-13,8) | | 分子名称: | capsid protein | | 著者 | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | 登録日 | 2019-08-15 | | 公開日 | 2020-08-26 | | 最終更新日 | 2022-03-30 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
