9IHX
 
 | |
9IHV
 
 | |
9IHT
 
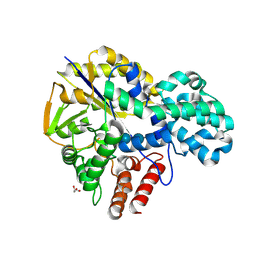 | | Crystal structure of GH57 family amylopullulanase from Aquifex aeolicus in complex with acarbose | | 分子名称: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, GLYCEROL, Glycoside hydrolase family 57 N-terminal domain-containing protein | | 著者 | Zhu, Z.M, Wang, W.W, Yu, F. | | 登録日 | 2024-06-18 | | 公開日 | 2025-06-04 | | 実験手法 | X-RAY DIFFRACTION (1.692 Å) | | 主引用文献 | The crystal structure of GH57 family amylopullulanase reveals its dual binding pockets sharing the same catalytic dyad.
Commun Biol, 8, 2025
|
|
8Y9W
 
 | |
7WIG
 
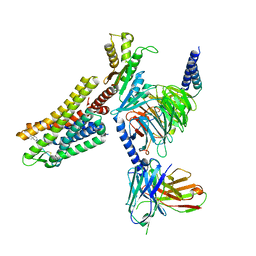 | | Cryo-EM structure of the L-054,264-bound human SSTR2-Gi1 complex | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | 著者 | Chen, L, Wang, W, Dong, Y, Shen, D, Guo, J, Qin, J, Zhang, H, Shen, Q, Zhang, Y, Mao, C. | | 登録日 | 2022-01-03 | | 公開日 | 2022-06-01 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Structures of the endogenous peptide- and selective non-peptide agonist-bound SSTR2 signaling complexes.
Cell Res., 32, 2022
|
|
7WIC
 
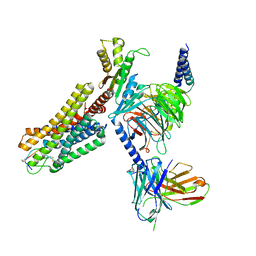 | | Cryo-EM structure of the SS-14-bound human SSTR2-Gi1 complex | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | 著者 | Chen, L, Wang, W, Dong, Y, Shen, D, Guo, J, Qin, J, Zhang, H, Shen, Q, Zhang, Y, Mao, C. | | 登録日 | 2022-01-03 | | 公開日 | 2022-06-01 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structures of the endogenous peptide- and selective non-peptide agonist-bound SSTR2 signaling complexes.
Cell Res., 32, 2022
|
|
6MIN
 
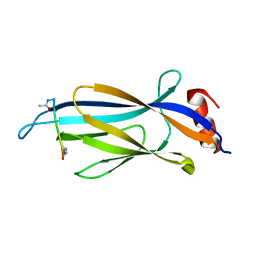 | |
6MIM
 
 | |
6MIQ
 
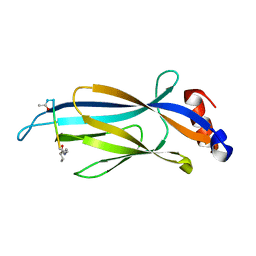 | | Crystal structure of Taf14 YEATS domain in complex with histone H3K9bu | | 分子名称: | Histone H3K9bu, Transcription initiation factor TFIID subunit 14 | | 著者 | Klein, B.J, Andrews, F.H, Vann, K.R, Kutateladze, T.G. | | 登録日 | 2018-09-19 | | 公開日 | 2018-11-14 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural insights into the pi-pi-pi stacking mechanism and DNA-binding activity of the YEATS domain.
Nat Commun, 9, 2018
|
|
6MIO
 
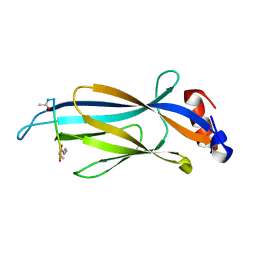 | | Crystal structure of Taf14 YEATS domain in complex with histone H3K9pr | | 分子名称: | Histone H3K9pr, Transcription initiation factor TFIID subunit 14 | | 著者 | Klein, B.J, Andrews, F.H, Vann, K.R, Kutateladze, T.G. | | 登録日 | 2018-09-19 | | 公開日 | 2018-11-14 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural insights into the pi-pi-pi stacking mechanism and DNA-binding activity of the YEATS domain.
Nat Commun, 9, 2018
|
|
6MIP
 
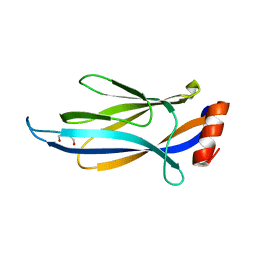 | |
6MIL
 
 | | Crystal structure of AF9 YEATS domain in complex with histone H3K9bu | | 分子名称: | DI(HYDROXYETHYL)ETHER, Histone H3K9bu, MALONATE ION, ... | | 著者 | Vann, K.R, Klein, B.J, Kutateladze, T.G. | | 登録日 | 2018-09-19 | | 公開日 | 2018-11-14 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Structural insights into the pi-pi-pi stacking mechanism and DNA-binding activity of the YEATS domain.
Nat Commun, 9, 2018
|
|
6AEO
 
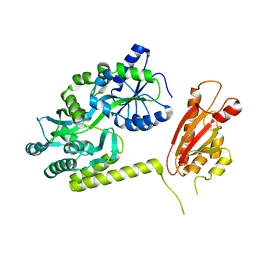 | | TssL periplasmic domain | | 分子名称: | GLYCEROL, Maltose/maltodextrin-binding periplasmic protein,TssL | | 著者 | Ran, T.T, Wang, W.W, Wang, X.B, Xu, D.Q. | | 登録日 | 2018-08-06 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of the periplasmic domain of TssL, a key membrane component of Type VI secretion system.
Int.J.Biol.Macromol., 120, 2018
|
|
2NX9
 
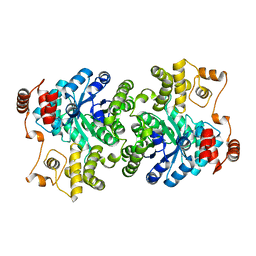 | |
8IKL
 
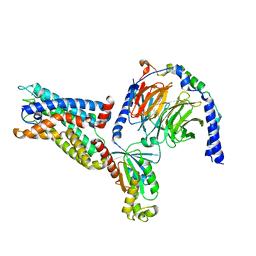 | | Cryo-EM structure of the CD97-G13 complex | | 分子名称: | Adhesion G protein-coupled receptor E5, Guanine nucleotide-binding protein G(13) subunit alpha isoforms short, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Mao, C, Zhao, R, Dong, Y, Gao, M, Chen, L, Zhang, C, Xiao, P. | | 登録日 | 2023-02-28 | | 公開日 | 2024-01-24 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (2.33 Å) | | 主引用文献 | Conformational transitions and activation of the adhesion receptor CD97.
Mol.Cell, 84, 2024
|
|
7Y86
 
 | | CcpS mutant | | 分子名称: | UPF0297 protein A7J08_00425 | | 著者 | Tang, J.S, Ran, T.T, Wang, W.W, Fan, H.J. | | 登録日 | 2022-06-22 | | 公開日 | 2023-05-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | A link between STK signalling and capsular polysaccharide synthesis in Streptococcus suis.
Nat Commun, 14, 2023
|
|
7Y8Z
 
 | | CcpS | | 分子名称: | UPF0297 protein A7J08_00425 | | 著者 | Tang, J.S, Ran, T.T, Wang, W.W, Fan, H.J. | | 登録日 | 2022-06-24 | | 公開日 | 2023-05-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | A link between STK signalling and capsular polysaccharide synthesis in Streptococcus suis.
Nat Commun, 14, 2023
|
|
6KJU
 
 | | Huge conformation shift of Vibrio cholerae VqmA dimer in the absence of target DNA provides insight into DNA-binding mechanisms of LuxR-type receptors | | 分子名称: | 3,5-dimethylpyrazin-2-ol, Helix-turn-helix transcriptional regulator | | 著者 | Wu, H, Li, M.J, Guo, H.J, Zhou, H, Wang, W.W, Xu, Q, Xu, C.Y, Yu, F, He, J.H. | | 登録日 | 2019-07-23 | | 公開日 | 2019-11-13 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Large conformation shifts of Vibrio cholerae VqmA dimer in the absence of target DNA provide insight into DNA-binding mechanisms of LuxR-type receptors.
Biochem.Biophys.Res.Commun., 520, 2019
|
|
8Y9V
 
 | | ZIKV NS2B/NS3 protease | | 分子名称: | DAR-LYS-ORN-ARG, Serine protease NS3, Serine protease subunit NS2B | | 著者 | Zhang, C.Y, Xu, Q, Wang, W.W, Zhou, H, Wang, Q.S. | | 登録日 | 2024-02-07 | | 公開日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystallographic data collection using a multilayer monochromator on an undulator beamline at SSRF
To Be Published
|
|
8Y4G
 
 | | Crystal structure of SARS-Cov-2 main protease G15S mutant in complex with Bofutrelvir | | 分子名称: | 3C-like proteinase nsp5, ~{N}-[(2~{S})-3-cyclohexyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | | 著者 | Zeng, P, Wang, W.W, Zhang, J, Li, J. | | 登録日 | 2024-01-30 | | 公開日 | 2025-01-08 | | 最終更新日 | 2025-04-09 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Inhibitory efficacy and structural insights of Bofutrelvir against SARS-CoV-2 M pro mutants and MERS-CoV M pro.
Commun Biol, 8, 2025
|
|
8Y4H
 
 | | Crystal structure of SARS-Cov-2 main protease K90R mutant in complex with Bofutrelvir | | 分子名称: | 3C-like proteinase nsp5, ~{N}-[(2~{S})-3-cyclohexyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | | 著者 | Guo, L, Wang, W.W, Zhang, J, Li, J. | | 登録日 | 2024-01-30 | | 公開日 | 2025-01-08 | | 最終更新日 | 2025-04-09 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Inhibitory efficacy and structural insights of Bofutrelvir against SARS-CoV-2 M pro mutants and MERS-CoV M pro.
Commun Biol, 8, 2025
|
|
8YWY
 
 | | Crystal structure of SARS-Cov-2 main protease E166N mutant in complex with Bofutrelvir | | 分子名称: | 3C-like proteinase nsp5, ~{N}-[(2~{S})-3-cyclohexyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | | 著者 | Zhou, X.L, Wang, W.W, Zhang, J, Li, J. | | 登録日 | 2024-04-01 | | 公開日 | 2025-04-09 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Inhibitory efficacy and structural insights of Bofutrelvir against SARS-CoV-2 M pro mutants and MERS-CoV M pro.
Commun Biol, 8, 2025
|
|
8YWZ
 
 | | Crystal structure of SARS-Cov-2 main protease H163A mutant in complex with Bofutrelvir | | 分子名称: | 3C-like proteinase nsp5, ~{N}-[(2~{S})-3-cyclohexyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | | 著者 | Zou, X.F, Wang, W.W, Zhang, J, Li, J. | | 登録日 | 2024-04-01 | | 公開日 | 2025-04-09 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Inhibitory efficacy and structural insights of Bofutrelvir against SARS-CoV-2 M pro mutants and MERS-CoV M pro.
Commun Biol, 8, 2025
|
|
