9EZ0
 
 | | Poliovirus type 1 (strain Mahoney) expanded conformation stabilised virus-like particle (PV1 SC6b) from a yeast expression system. | | 分子名称: | Capsid protein VP0, Capsid protein VP1, Capsid protein VP3 | | 著者 | Bahar, M.W, Sherry, L, Stonehouse, N.J, Rowlands, D.J, Fry, E.E, Stuart, D.I. | | 登録日 | 2024-04-10 | | 公開日 | 2025-01-29 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Recombinant expression systems for production of stabilised virus-like particles as next-generation polio vaccines.
Nat Commun, 16, 2025
|
|
9F0K
 
 | | Poliovirus type 1 (strain Mahoney) expanded conformation stabilised virus-like particle (PV1 SC6b) from a mammalian expression system | | 分子名称: | Capsid protein VP0, Capsid protein VP1, Capsid protein VP3 | | 著者 | Bahar, M.W, Porta, C, Fry, E.E, Stuart, D.I. | | 登録日 | 2024-04-16 | | 公開日 | 2025-01-29 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Recombinant expression systems for production of stabilised virus-like particles as next-generation polio vaccines.
Nat Commun, 16, 2025
|
|
9F3Q
 
 | | Poliovirus type 1 (strain Mahoney) stabilised virus-like particle (PV1 SC6b) in complex with GPP3 and GSH. | | 分子名称: | 1-[(3S)-5-[4-[(E)-ETHOXYIMINOMETHYL]PHENOXY]-3-METHYL-PENTYL]-3-PYRIDIN-4-YL-IMIDAZOLIDIN-2-ONE, Capsid protein VP0, Capsid protein VP1, ... | | 著者 | Bahar, M.W, Nasta, V, Sherry, L, Stonehouse, N.J, Rowlands, D.J, Fry, E.E, Stuart, D.I. | | 登録日 | 2024-04-25 | | 公開日 | 2025-01-29 | | 実験手法 | ELECTRON MICROSCOPY (2.75 Å) | | 主引用文献 | Recombinant expression systems for production of stabilised virus-like particles as next-generation polio vaccines.
Nat Commun, 16, 2025
|
|
9F5P
 
 | | Poliovirus type 2 (strain MEF-1) stabilised virus-like particle (PV2 SC6b) from an insect cell expression system. | | 分子名称: | Capsid protein VP0, Capsid protein VP1, Capsid protein VP3, ... | | 著者 | Bahar, M.W, Porta, C, Fry, E.E, Stuart, D.I. | | 登録日 | 2024-04-29 | | 公開日 | 2025-01-29 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Recombinant expression systems for production of stabilised virus-like particles as next-generation polio vaccines.
Nat Commun, 16, 2025
|
|
9EYY
 
 | | Poliovirus type 1 (strain Mahoney) native conformation stabilised virus-like particle (PV1 SC6b) from a yeast expression system. | | 分子名称: | Capsid protein VP0, Capsid protein VP1, Capsid protein VP3, ... | | 著者 | Bahar, M.W, Sherry, L, Stonehouse, N.J, Rowlands, D.J, Fry, E.E, Stuart, D.I. | | 登録日 | 2024-04-09 | | 公開日 | 2025-01-29 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Recombinant expression systems for production of stabilised virus-like particles as next-generation polio vaccines.
Nat Commun, 16, 2025
|
|
3KYJ
 
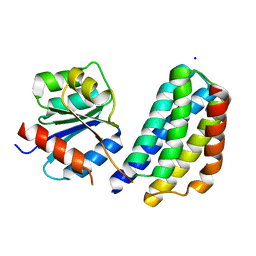 | | Crystal structure of the P1 domain of CheA3 in complex with CheY6 from R. sphaeroides | | 分子名称: | CheY6 protein, Putative histidine protein kinase, SODIUM ION | | 著者 | Bell, C.H, Porter, S.L, Armitage, J.P, Stuart, D.I. | | 登録日 | 2009-12-06 | | 公開日 | 2010-02-16 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Using structural information to change the phosphotransfer specificity of a two-component chemotaxis signalling complex
Plos Biol., 8, 2010
|
|
3ME4
 
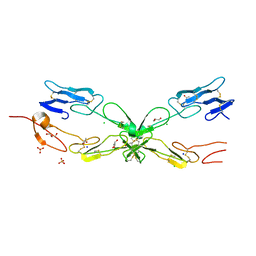 | | Crystal structure of mouse RANK | | 分子名称: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | 著者 | Walter, S.W, Liu, C, Zhu, X, Wu, Y, Owens, R.J, Stuart, D.I, Gao, B, Ren, J. | | 登録日 | 2010-03-31 | | 公開日 | 2010-06-02 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Structural and Functional Insights of RANKL-RANK Interaction and Signaling.
J.Immunol., 2010
|
|
3ME2
 
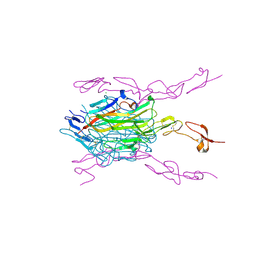 | | Crystal structure of mouse RANKL-RANK complex | | 分子名称: | CHLORIDE ION, SODIUM ION, Tumor necrosis factor ligand superfamily member 11, ... | | 著者 | Walter, S.W, Liu, C.Z, Zhu, X.K, Wu, Y, Owens, R.J, Stuart, D.I, Gao, B, Ren, J. | | 登録日 | 2010-03-31 | | 公開日 | 2010-06-02 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural and Functional Insights of RANKL-RANK Interaction and Signaling.
J.Immunol., 2010
|
|
8BS8
 
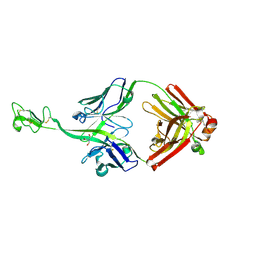 | | Bovine naive ultralong antibody AbD08 collected at 100K | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Heavy chain, Light chain | | 著者 | Clarke, J.D, Douangamath, A, Mikolajek, H, Stuart, D.I, Owens, R.J. | | 登録日 | 2022-11-24 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | The impact of exchanging the light and heavy chains on the structures of bovine ultralong antibodies.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
3KYI
 
 | | Crystal structure of the phosphorylated P1 domain of CheA3 in complex with CheY6 from R. sphaeroides | | 分子名称: | CheY6 protein, Putative histidine protein kinase | | 著者 | Bell, C.H, Porter, S.L, Armitage, J.P, Stuart, D.I. | | 登録日 | 2009-12-06 | | 公開日 | 2010-02-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Using structural information to change the phosphotransfer specificity of a two-component chemotaxis signalling complex
Plos Biol., 8, 2010
|
|
9F9Y
 
 | | SARS-CoV-2 BA-2.87.1 Spike ectodomain | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | 著者 | Ren, J, Stuart, D.I, Duyvesteyn, H.M.E. | | 登録日 | 2024-05-09 | | 公開日 | 2024-08-21 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Concerted deletions eliminate a neutralizing supersite in SARS-CoV-2 BA.2.87.1 spike.
Structure, 32, 2024
|
|
3DUZ
 
 | | Crystal structure of the postfusion form of baculovirus fusion protein GP64 | | 分子名称: | MERCURY (II) ION, Major envelope glycoprotein | | 著者 | Kadlec, J, Loureiro, S, Abrescia, N.G.A, Jones, I.M, Stuart, D.I. | | 登録日 | 2008-07-18 | | 公開日 | 2008-09-16 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | The postfusion structure of baculovirus gp64 supports a unified view of viral fusion machines.
Nat.Struct.Mol.Biol., 15, 2008
|
|
1ALC
 
 | |
1HML
 
 | | ALPHA_LACTALBUMIN POSSESSES A DISTINCT ZINC BINDING SITE | | 分子名称: | ALPHA-LACTALBUMIN, CALCIUM ION, SULFATE ION, ... | | 著者 | Ren, J, Stuart, D.I, Acharya, K.R. | | 登録日 | 1994-09-29 | | 公開日 | 1995-01-26 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Alpha-lactalbumin possesses a distinct zinc binding site.
J.Biol.Chem., 268, 1993
|
|
1HNF
 
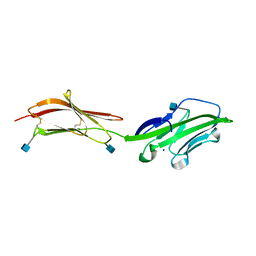 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR REGION OF THE HUMAN CELL ADHESION MOLECULE CD2 AT 2.5 ANGSTROMS RESOLUTION | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CD2, SODIUM ION | | 著者 | Bodian, D.L, Jones, E.Y, Harlos, K, Stuart, D.I, Davis, S.J. | | 登録日 | 1994-08-10 | | 公開日 | 1995-02-07 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of the extracellular region of the human cell adhesion molecule CD2 at 2.5 A resolution.
Structure, 2, 1994
|
|
1HNG
 
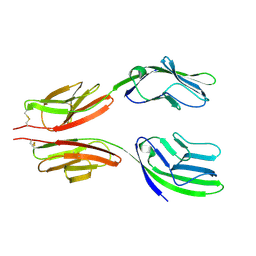 | | CRYSTAL STRUCTURE AT 2.8 ANGSTROMS RESOLUTION OF A SOLUBLE FORM OF THE CELL ADHESION MOLECULE CD2 | | 分子名称: | CD2 | | 著者 | Jones, E.Y, Davis, S.J, Williams, A.F, Harlos, K, Stuart, D.I. | | 登録日 | 1994-08-10 | | 公開日 | 1995-02-07 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure at 2.8 A resolution of a soluble form of the cell adhesion molecule CD2.
Nature, 360, 1992
|
|
5WTG
 
 | | Crystal structure of the Fab fragment of anti-HAV antibody R10 | | 分子名称: | FAB Heavy chain, FAB Light chain | | 著者 | Wang, X, Zhu, L, Dang, M, Hu, Z, Gao, Q, Yuan, S, Sun, Y, Zhang, B, Ren, J, Walter, T.S, Wang, J, Fry, E.E, Stuart, D.I, Rao, Z. | | 登録日 | 2016-12-11 | | 公開日 | 2017-01-25 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.907 Å) | | 主引用文献 | Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5WTE
 
 | | Cryo-EM structure for Hepatitis A virus full particle | | 分子名称: | VP1, VP2, VP3 | | 著者 | Wang, X, Zhu, L, Dang, M, Hu, Z, Gao, Q, Yuan, S, Sun, Y, Zhang, B, Ren, J, Walter, T.S, Wang, J, Fry, E.E, Stuart, D.I, Rao, Z. | | 登録日 | 2016-12-11 | | 公開日 | 2017-01-25 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5WTH
 
 | | Cryo-EM structure for Hepatitis A virus complexed with FAB | | 分子名称: | FAB Heavy Chain, FAB Light Chain, Polyprotein, ... | | 著者 | Wang, X, Zhu, L, Dang, M, Hu, Z, Gao, Q, Yuan, S, Sun, Y, Zhang, B, Ren, J, Walter, T.S, Wang, J, Fry, E.E, Stuart, D.I, Rao, Z. | | 登録日 | 2016-12-12 | | 公開日 | 2017-01-25 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5WTF
 
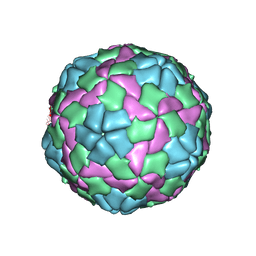 | | Cryo-EM structure for Hepatitis A virus empty particle | | 分子名称: | VP0, VP1, VP3 | | 著者 | Wang, X, Zhu, L, Dang, M, Hu, Z, Gao, Q, Yuan, S, Sun, Y, Zhang, B, Ren, J, Walter, T.S, Wang, J, Fry, E.E, Stuart, D.I, Rao, Z. | | 登録日 | 2016-12-11 | | 公開日 | 2017-01-25 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1MHE
 
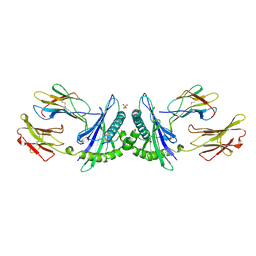 | | THE HUMAN NON-CLASSICAL MAJOR HISTOCOMPATIBILITY COMPLEX MOLECULE HLA-E | | 分子名称: | BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN HLA-E, PEPTIDE (VMAPRTVLL), ... | | 著者 | O'Callaghan, C.A, Tormo, J, Willcox, B.E, Braud, V.B, Jakobsen, B.K, Stuart, D.I, Mcmichael, A.J, Bell, J.I, Jones, E.Y. | | 登録日 | 1998-08-24 | | 公開日 | 1999-03-23 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Structural features impose tight peptide binding specificity in the nonclassical MHC molecule HLA-E.
Mol.Cell, 1, 1998
|
|
1MU2
 
 | | CRYSTAL STRUCTURE OF HIV-2 REVERSE TRANSCRIPTASE | | 分子名称: | GLYCEROL, HIV-2 RT, SULFATE ION | | 著者 | Ren, J, Bird, L.E, Chamberlain, P.P, Stewart-Jones, G.B, Stuart, D.I, Stammers, D.K. | | 登録日 | 2002-09-23 | | 公開日 | 2002-10-30 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structure of HIV-2 reverse transcriptase at 2.35-A resolution and the mechanism of resistance to non-nucleoside inhibitors
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
5MQW
 
 | | High-speed fixed-target serial virus crystallography | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Roedig, P, Ginn, H.M, Pakendorf, T, Sutton, G, Harlos, K, Walter, T.S, Meyer, J, Fischer, P, Duman, R, Vartiainen, I, Reime, B, Warmer, M, Brewster, A.S, Young, I.D, Michels-Clark, T, Sauter, N.K, Sikorsky, M, Nelson, S, Damiani, D.S, Alonso-Mori, R, Ren, J, Fry, E.E, David, C, Stuart, D.I, Wagner, A, Meents, A. | | 登録日 | 2016-12-21 | | 公開日 | 2017-06-21 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | High-speed fixed-target serial virus crystallography.
Nat. Methods, 14, 2017
|
|
4FJV
 
 | | Crystal Structure of Human Otubain2 and Ubiquitin Complex | | 分子名称: | ETHANAMINE, GLYCEROL, Polyubiquitin-C, ... | | 著者 | Altun, M, Walter, T.S, Kramer, H.B, Iphofer, A, David, Y, Komsany, A, Ternette, N, Nicholson, B, Navon, A, Stuart, D.I, Ren, J, Kessler, B.M. | | 登録日 | 2012-06-12 | | 公開日 | 2013-06-12 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.047 Å) | | 主引用文献 | The human otubain2-ubiquitin structure provides insights into the cleavage specificity of poly-ubiquitin-linkages.
Plos One, 10, 2015
|
|
1BBT
 
 | | METHODS USED IN THE STRUCTURE DETERMINATION OF FOOT AND MOUTH DISEASE VIRUS | | 分子名称: | FOOT-AND-MOUTH DISEASE VIRUS (SUBUNIT VP1), FOOT-AND-MOUTH DISEASE VIRUS (SUBUNIT VP2), FOOT-AND-MOUTH DISEASE VIRUS (SUBUNIT VP3), ... | | 著者 | Acharya, K.R, Fry, E.E, Logan, D.T, Stuart, D.I. | | 登録日 | 1992-05-18 | | 公開日 | 1994-01-31 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Methods used in the structure determination of foot-and-mouth disease virus.
Acta Crystallogr.,Sect.A, 49, 1993
|
|
