5YZW
 
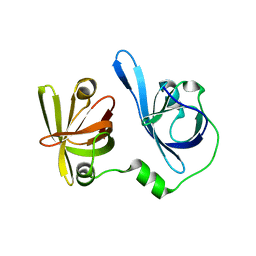 | | Crystal structure of p204 HINb domain | | 分子名称: | Ifi204 | | 著者 | Jin, T. | | 登録日 | 2017-12-15 | | 公開日 | 2018-12-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.001 Å) | | 主引用文献 | Structural mechanism of DNA recognition by the p204 HIN domain.
Nucleic Acids Res., 2021
|
|
5Z7D
 
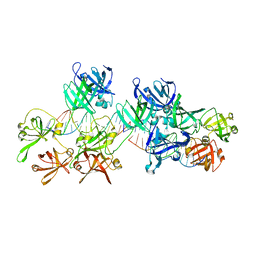 | | p204HINab-dsDNA complex structure | | 分子名称: | DNA (5'-D(P*CP*CP*AP*TP*CP*AP*GP*AP*AP*AP*GP*AP*GP*AP*GP*C)-3'), Interferon-activable protein 204 | | 著者 | Jin, T, Jiang, J, Xiao, T.S. | | 登録日 | 2018-01-28 | | 公開日 | 2019-01-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (4.5 Å) | | 主引用文献 | Structural mechanism of DNA recognition by the p204 HIN domain.
Nucleic Acids Res., 2021
|
|
5H7Q
 
 | |
5H7N
 
 | |
5WQ6
 
 | |
5WPZ
 
 | |
5E7P
 
 | | Crystal Structure of MSMEG_0858 (Uniprot A0QQS4), a AAA ATPase. | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein Cdc48, GLYCEROL, ... | | 著者 | Unciulac-Carp, M, Smith, P, Shuman, S. | | 登録日 | 2015-10-12 | | 公開日 | 2016-08-24 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.507 Å) | | 主引用文献 | Crystal Structure and Biochemical Characterization of a Mycobacterium smegmatis AAA-Type Nucleoside Triphosphatase Phosphohydrolase (Msm0858).
J.Bacteriol., 198, 2016
|
|
1CRL
 
 | |
4DOM
 
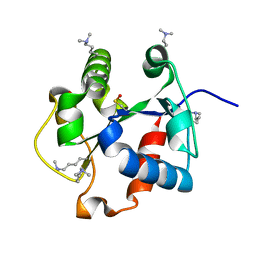 | |
1LPM
 
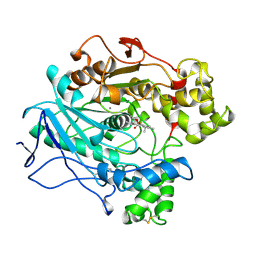 | | A STRUCTURAL BASIS FOR THE CHIRAL PREFERENCES OF LIPASES | | 分子名称: | (1R)-MENTHYL HEXYL PHOSPHONATE GROUP, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Grochulski, P.G, Cygler, M.C. | | 登録日 | 1995-01-06 | | 公開日 | 1995-04-20 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | A Structural Basis for the Chiral Preferences of Lipases
J.Am.Chem.Soc., 116, 1994
|
|
1LPS
 
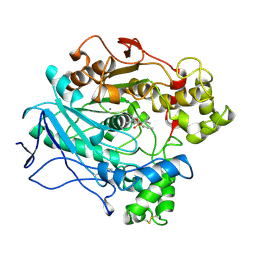 | | A STRUCTURAL BASIS FOR THE CHIRAL PREFERENCES OF LIPASES | | 分子名称: | (1S)-MENTHYL HEXYL PHOSPHONATE GROUP, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Grochulski, P.G, Cygler, M.C. | | 登録日 | 1995-01-05 | | 公開日 | 1995-02-14 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | A Structural Basis for the Chiral Preferences of Lipases
J.Am.Chem.Soc., 116, 1994
|
|
1LPN
 
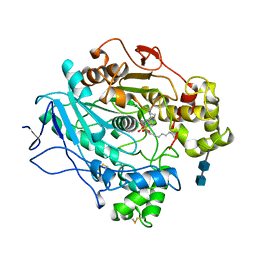 | |
1LPP
 
 | |
1LPO
 
 | |
1TRH
 
 | |
3B6Z
 
 | | Lovastatin polyketide enoyl reductase (LovC) complexed with 2'-phosphoadenosyl isomer of crotonoyl-CoA | | 分子名称: | Enoyl reductase, GLYCEROL, S-{(9R,13R,15S)-17-[(2R,3R,4R,5R)-5-(6-amino-9H-purin-9-yl)-3-hydroxy-4-(phosphonooxy)tetrahydrofuran-2-yl]-9,13,15-trihydroxy-10,10-dimethyl-13,15-dioxido-4,8-dioxo-12,14,16-trioxa-3,7-diaza-13,15-diphosphaheptadec-1-yl}(2E)-but-2-enethioate | | 著者 | Ames, B.D, Smith, P.T, Ma, S.M, Wong, E.W, Xie, X, Vederas, J.C, Tang, Y, Tsai, S.-C. | | 登録日 | 2007-10-29 | | 公開日 | 2008-09-16 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Crystal structure and biochemical studies of the trans-acting polyketide enoyl reductase LovC from lovastatin biosynthesis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3B70
 
 | | Crystal structure of Aspergillus terreus trans-acting lovastatin polyketide enoyl reductase (LovC) with bound NADP | | 分子名称: | Enoyl reductase, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Ames, B.D, Smith, P.T, Ma, S.M, Wong, E.W, Xie, X, Vederas, J.C, Tang, Y, Tsai, S.-C. | | 登録日 | 2007-10-29 | | 公開日 | 2008-09-16 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Crystal structure and biochemical studies of the trans-acting polyketide enoyl reductase LovC from lovastatin biosynthesis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2MN3
 
 | | Structure of Platypus 'Intermediate' Defensin-like Peptide (Int-DLP) | | 分子名称: | Defensin-BvL | | 著者 | Torres, A.M, Bansal, P.S, Koh, J.M.S, Pages, G, Wu, M.J, Kuchel, P.W. | | 登録日 | 2014-03-27 | | 公開日 | 2014-09-17 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure and antimicrobial activity of platypus 'intermediate' defensin-like peptide.
Febs Lett., 588, 2014
|
|
3RN5
 
 | | Structural basis of cytosolic DNA recognition by innate immune receptors | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*TP*CP*AP*AP*AP*GP*AP*GP*AP*GP*AP*AP*AP*GP*AP*G)-3'), DNA (5'-D(*GP*CP*TP*CP*TP*TP*TP*CP*TP*CP*TP*CP*TP*TP*TP*GP*AP*TP*G)-3'), ... | | 著者 | Jin, T.C, Xiao, T. | | 登録日 | 2011-04-21 | | 公開日 | 2012-04-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structures of the HIN Domain:DNA Complexes Reveal Ligand Binding and Activation Mechanisms of the AIM2 Inflammasome and IFI16 Receptor.
Immunity, 36, 2012
|
|
3RNU
 
 | | Structural Basis of Cytosolic DNA Sensing by Innate Immune Receptors | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*CP*AP*TP*CP*AP*AP*AP*GP*AP*GP*AP*GP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*CP*TP*CP*TP*TP*TP*GP*AP*TP*GP*GP*CP*C)-3'), ... | | 著者 | Jin, T.C, Xiao, T. | | 登録日 | 2011-04-22 | | 公開日 | 2012-04-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.502 Å) | | 主引用文献 | Structures of the HIN Domain:DNA Complexes Reveal Ligand Binding and Activation Mechanisms of the AIM2 Inflammasome and IFI16 Receptor.
Immunity, 36, 2012
|
|
3RN2
 
 | | Structural Basis of Cytosolic DNA Recognition by Innate Immune Receptors | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*TP*CP*AP*AP*AP*GP*AP*TP*CP*TP*TP*TP*GP*AP*TP*GP*G)-3'), Interferon-inducible protein AIM2 | | 著者 | Jin, T.C, Xiao, T. | | 登録日 | 2011-04-21 | | 公開日 | 2012-04-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structures of the HIN Domain:DNA Complexes Reveal Ligand Binding and Activation Mechanisms of the AIM2 Inflammasome and IFI16 Receptor.
Immunity, 36, 2012
|
|
3RLN
 
 | |
3RLO
 
 | |
2REZ
 
 | | Tetracenomycin ARO/CYC NaI Structure | | 分子名称: | ACETATE ION, IODIDE ION, Multifunctional cyclase-dehydratase-3-O-methyl transferase tcmN | | 著者 | Ames, B.D, Tsai, S.C. | | 登録日 | 2007-09-27 | | 公開日 | 2008-04-22 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure and functional analysis of tetracenomycin ARO/CYC: implications for cyclization specificity of aromatic polyketides.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2RER
 
 | |
