6IXV
 
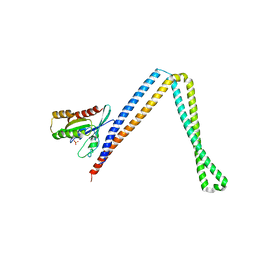 | | Crystal structure of SH3BP5-Rab11a | | 分子名称: | PHOSPHATE ION, Ras-related protein Rab-11A, SH3 domain-binding protein 5 | | 著者 | Goto-Ito, S, Yamagata, A, Sato, Y, Fukai, S. | | 登録日 | 2018-12-12 | | 公開日 | 2019-03-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Structural basis of guanine nucleotide exchange for Rab11 by SH3BP5.
Life Sci Alliance, 2, 2019
|
|
6IXG
 
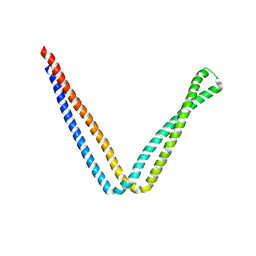 | |
6IXE
 
 | | Crystal structure of SeMet apo SH3BP5 (I41) | | 分子名称: | SH3 domain-binding protein 5, SUCCINIC ACID | | 著者 | Goto-Ito, S, Yamagata, A, Sato, Y, Fukai, S. | | 登録日 | 2018-12-10 | | 公開日 | 2019-03-20 | | 最終更新日 | 2019-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.35 Å) | | 主引用文献 | Structural basis of guanine nucleotide exchange for Rab11 by SH3BP5.
Life Sci Alliance, 2, 2019
|
|
5XNZ
 
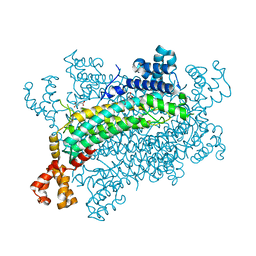 | | Crystal structure of CreD complex with fumarate | | 分子名称: | CreD, FUMARIC ACID | | 著者 | Katsuyama, Y, Sato, Y, Sugai, Y, Higashiyama, Y, Senda, M, Senda, T, Ohnishi, Y. | | 登録日 | 2017-05-25 | | 公開日 | 2018-03-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of the nitrosuccinate lyase CreD in complex with fumarate provides insights into the catalytic mechanism for nitrous acid elimination
FEBS J., 285, 2018
|
|
5XNY
 
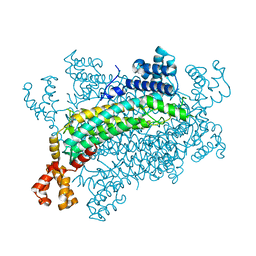 | | Crystal structure of CreD | | 分子名称: | CreD | | 著者 | Katsuyama, Y, Sato, Y, Sugai, Y, Higashiyama, Y, Senda, M, Senda, T, Ohnishi, Y. | | 登録日 | 2017-05-25 | | 公開日 | 2018-03-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Crystal structure of the nitrosuccinate lyase CreD in complex with fumarate provides insights into the catalytic mechanism for nitrous acid elimination
FEBS J., 285, 2018
|
|
3A58
 
 | | Crystal structure of Sec3p - Rho1p complex from Saccharomyces cerevisiae | | 分子名称: | Exocyst complex component SEC3, GTP-binding protein RHO1, MAGNESIUM ION, ... | | 著者 | Yamashita, M, Sato, Y, Yamagata, A, Mimura, H, Yoshikawa, A, Fukai, S. | | 登録日 | 2009-08-03 | | 公開日 | 2010-01-12 | | 最終更新日 | 2021-11-10 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis for the Rho- and phosphoinositide-dependent localization of the exocyst subunit Sec3
Nat.Struct.Mol.Biol., 17, 2010
|
|
5XIT
 
 | | Crystal structure of RNF168 UDM1 in complex with Lys63-linked diubiquitin, form II | | 分子名称: | E3 ubiquitin-protein ligase RNF168, GLYCEROL, PRASEODYMIUM ION, ... | | 著者 | Takahashi, T.S, Sato, Y, Fukai, S. | | 登録日 | 2017-04-27 | | 公開日 | 2018-03-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structural insights into two distinct binding modules for Lys63-linked polyubiquitin chains in RNF168.
Nat Commun, 9, 2018
|
|
5XIS
 
 | | Crystal structure of RNF168 UDM1 in complex with Lys63-linked diubiquitin, form I | | 分子名称: | E3 ubiquitin-protein ligase RNF168, MAGNESIUM ION, Ubiquitin-40S ribosomal protein S27a, ... | | 著者 | Takahashi, T.S, Sato, Y, Fukai, S. | | 登録日 | 2017-04-27 | | 公開日 | 2018-03-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Structural insights into two distinct binding modules for Lys63-linked polyubiquitin chains in RNF168.
Nat Commun, 9, 2018
|
|
5YDK
 
 | | Crystal structure of RNF168 UDM1 in complex with Lys63-linked diubiquitin, tetrameric form | | 分子名称: | E3 ubiquitin-protein ligase RNF168, GLYCEROL, Ubiquitin-40S ribosomal protein S27a | | 著者 | Takahashi, T.S, Sato, Y, Fukai, S. | | 登録日 | 2017-09-13 | | 公開日 | 2018-03-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.505 Å) | | 主引用文献 | Structural insights into two distinct binding modules for Lys63-linked polyubiquitin chains in RNF168
Nat Commun, 9, 2018
|
|
5YOY
 
 | | Crystal structure of the human tumor necrosis factor in complex with golimumab Fv | | 分子名称: | Golimumab heavy chain variable region, Golimumab light chain variable region, Tumor necrosis factor | | 著者 | Ono, M, Horita, S, Sato, Y, Nomura, Y, Iwata, S, Nomura, N. | | 登録日 | 2017-10-31 | | 公開日 | 2018-05-09 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.727 Å) | | 主引用文献 | Structural basis for tumor necrosis factor blockade with the therapeutic antibody golimumab
Protein Sci., 27, 2018
|
|
5YJ9
 
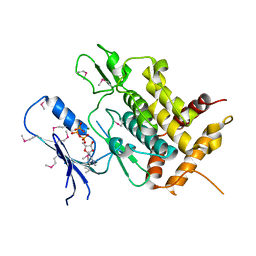 | | Crystal structure of Tribolium castaneum PINK1 kinase domain in complex with AMP-PNP | | 分子名称: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase PINK1, ... | | 著者 | Okatsu, K, Sato, Y, Fukai, S. | | 登録日 | 2017-10-09 | | 公開日 | 2018-07-25 | | 実験手法 | X-RAY DIFFRACTION (2.53 Å) | | 主引用文献 | Structural insights into ubiquitin phosphorylation by PINK1.
Sci Rep, 8, 2018
|
|
5XIU
 
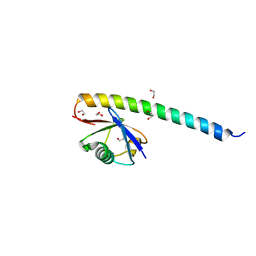 | | Crystal structure of RNF168 UDM2 in complex with Lys63-linked diubiquitin | | 分子名称: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase RNF168, Ubiquitin-40S ribosomal protein S27a | | 著者 | Takahashi, T.S, Sato, Y, Fukai, S. | | 登録日 | 2017-04-27 | | 公開日 | 2018-03-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural insights into two distinct binding modules for Lys63-linked polyubiquitin chains in RNF168.
Nat Commun, 9, 2018
|
|
1EHL
 
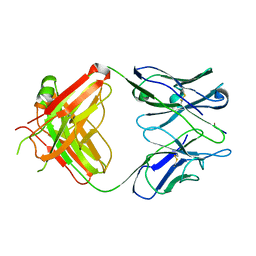 | | 64M-2 ANTIBODY FAB COMPLEXED WITH D(5HT)(6-4)T | | 分子名称: | 5'-(D(5HT)P*(6-4)T)-3', ANTI-(6-4) PHOTOPRODUCT ANTIBODY 64M-2 FAB (HEAVY CHAIN), ANTI-(6-4) PHOTOPRODUCT ANTIBODY 64M-2 FAB (LIGHT CHAIN) | | 著者 | Yokoyama, H, Mizutani, R, Satow, Y, Komatsu, Y, Ohtsuka, E, Nikaido, O. | | 登録日 | 2000-02-21 | | 公開日 | 2001-02-21 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of the 64M-2 antibody Fab fragment in complex with a DNA dT(6-4)T photoproduct formed by ultraviolet radiation.
J.Mol.Biol., 299, 2000
|
|
5ZNM
 
 | | Colicin D Central Domain and C-terminal tRNase domain | | 分子名称: | Colicin-D, GLYCEROL, SULFATE ION | | 著者 | Chang, J.W, Sato, Y, Ogawa, T, Arakawa, T, Fukai, S, Fushinobu, S, Masaki, H. | | 登録日 | 2018-04-10 | | 公開日 | 2018-08-29 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal structure of the central and the C-terminal RNase domains of colicin D implicated its translocation pathway through inner membrane of target cell
J. Biochem., 164, 2018
|
|
3A31
 
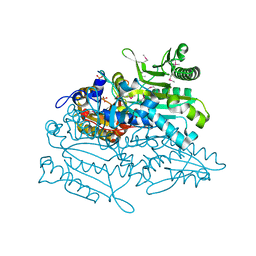 | | Crystal structure of putative threonyl-tRNA synthetase ThrRS-1 from Aeropyrum pernix (selenomethionine derivative) | | 分子名称: | Probable threonyl-tRNA synthetase 1, SULFATE ION, ZINC ION | | 著者 | Shimizu, S, Juan, E.C.M, Miyashita, Y, Sato, Y, Hoque, M.M, Suzuki, K, Yogiashi, M, Tsunoda, M, Dock-Bregeon, A.-C, Moras, D, Sekiguchi, T, Takenaka, A. | | 登録日 | 2009-06-07 | | 公開日 | 2009-10-27 | | 最終更新日 | 2013-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Two complementary enzymes for threonylation of tRNA in crenarchaeota: crystal structure of Aeropyrum pernix threonyl-tRNA synthetase lacking a cis-editing domain
J.Mol.Biol., 394, 2009
|
|
3A32
 
 | | Crystal structure of putative threonyl-tRNA synthetase ThrRS-1 from Aeropyrum pernix | | 分子名称: | Probable threonyl-tRNA synthetase 1, SULFATE ION, ZINC ION | | 著者 | Shimizu, S, Juan, E.C.M, Miyashita, Y, Sato, Y, Hoque, M.M, Suzuki, K, Yogiashi, M, Tsunoda, M, Dock-Bregeon, A.-C, Moras, D, Sekiguchi, T, Takenaka, A. | | 登録日 | 2009-06-07 | | 公開日 | 2009-10-27 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Two complementary enzymes for threonylation of tRNA in crenarchaeota: crystal structure of Aeropyrum pernix threonyl-tRNA synthetase lacking a cis-editing domain
J.Mol.Biol., 394, 2009
|
|
5XWU
 
 | | Crystal structure of PTPdelta Ig1-Ig3 in complex with SALM2 LRR-Ig | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Goto-Ito, S, Yamagata, A, Sato, Y, Fukai, S. | | 登録日 | 2017-06-30 | | 公開日 | 2018-06-06 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.162 Å) | | 主引用文献 | Structural basis of trans-synaptic interactions between PTP delta and SALMs for inducing synapse formation.
Nat Commun, 9, 2018
|
|
5XWT
 
 | | Crystal structure of PTPdelta Ig1-Fn1 in complex with SALM5 LRR-Ig | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Goto-Ito, S, Yamagata, A, Sato, Y, Fukai, S. | | 登録日 | 2017-06-30 | | 公開日 | 2018-06-06 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (4.178 Å) | | 主引用文献 | Structural basis of trans-synaptic interactions between PTP delta and SALMs for inducing synapse formation.
Nat Commun, 9, 2018
|
|
5XWS
 
 | | Crystal structure of SALM5 LRR-Ig | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Leucine-rich repeat and fibronectin type-III domain-containing protein 5 | | 著者 | Goto-Ito, S, Yamagata, A, Sato, Y, Fukai, S. | | 登録日 | 2017-06-30 | | 公開日 | 2018-06-06 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.084 Å) | | 主引用文献 | Structural basis of trans-synaptic interactions between PTP delta and SALMs for inducing synapse formation.
Nat Commun, 9, 2018
|
|
6A6I
 
 | | Crystal structure of the winged-helix domain of Cockayne syndrome group B protein in complex with ubiquitin | | 分子名称: | CHLORIDE ION, Excision repair cross-complementing rodent repair deficiency, complementation group 6 variant, ... | | 著者 | Takahashi, T.S, Sato, Y, Fukai, S. | | 登録日 | 2018-06-28 | | 公開日 | 2019-02-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis of ubiquitin recognition by the winged-helix domain of Cockayne syndrome group B protein.
Nucleic Acids Res., 47, 2019
|
|
3A36
 
 | | Structural insight into the membrane insertion of tail-anchored proteins by Get3 | | 分子名称: | ATPase GET3, ZINC ION | | 著者 | Yamagata, A, Mimura, H, Sato, Y, Yamashita, M, Yoshikawa, A, Fukai, S. | | 登録日 | 2009-06-10 | | 公開日 | 2010-01-12 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural insight into the membrane insertion of tail-anchored proteins by Get3
Genes Cells, 15, 2010
|
|
3A1K
 
 | | Crystal structure of Rhodococcus sp. N771 Amidase | | 分子名称: | Amidase | | 著者 | Ohtaki, A, Noguchi, K, Sato, Y, Murata, K, Odaka, M, Yohda, M. | | 登録日 | 2009-04-09 | | 公開日 | 2009-11-03 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Structure and characterization of amidase from Rhodococcus sp. N-771: Insight into the molecular mechanism of substrate recognition
Biochim.Biophys.Acta, 1804, 2010
|
|
2Z6E
 
 | | Crystal Structure of Human DAAM1 FH2 | | 分子名称: | Disheveled-associated activator of morphogenesis 1 | | 著者 | Yamashita, M, Higashi, T, Sato, Y, Shirakawa, R, Kita, T, Horiuchi, H, Fukai, S, Nureki, O. | | 登録日 | 2007-07-31 | | 公開日 | 2008-05-27 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of human DAAM1 formin homology 2 domain
Genes Cells, 12, 2007
|
|
3A1I
 
 | | Crystal structure of Rhodococcus sp. N-771 Amidase complexed with Benzamide | | 分子名称: | Amidase, BENZAMIDE | | 著者 | Ohtaki, A, Noguchi, K, Sato, Y, Murata, K, Odaka, M, Yohda, M. | | 登録日 | 2009-04-03 | | 公開日 | 2009-10-27 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Structure and Characterization of Amidase from Rhodococcus sp. N-771: Insight into the Molecular Mechanism of Substrate Recognition
Biochim.Biophys.Acta, 2009
|
|
3A37
 
 | | Structural insight into the membrane insertion of tail-anchored proteins by Get3 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATPase GET3, ZINC ION | | 著者 | Yamagata, A, Mimura, H, Sato, Y, Yamashita, M, Yoshikawa, A, Fukai, S. | | 登録日 | 2009-06-10 | | 公開日 | 2010-01-12 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural insight into the membrane insertion of tail-anchored proteins by Get3
Genes Cells, 15, 2010
|
|
