8IOT
 
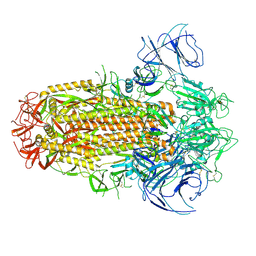 | | Structure of the SARS-CoV-2 XBB.1 spike glycoprotein (closed-2 state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Anraku, Y, Kita, S, Yajima, H, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | 登録日 | 2023-03-13 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (2.51 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants.
Nat Commun, 14, 2023
|
|
8IOU
 
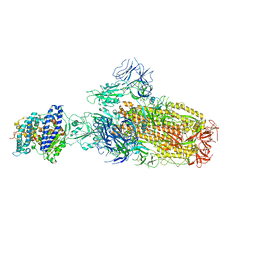 | | Structure of SARS-CoV-2 XBB.1 spike glycoprotein in complex with ACE2 (1-up state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Anraku, Y, Kita, S, Yajima, H, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | 登録日 | 2023-03-13 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants.
Nat Commun, 14, 2023
|
|
8IOS
 
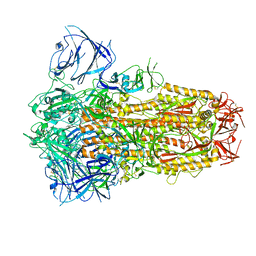 | | Structure of the SARS-CoV-2 XBB.1 spike glycoprotein (closed-1 state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Anraku, Y, Kita, S, Yajima, H, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | 登録日 | 2023-03-13 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants.
Nat Commun, 14, 2023
|
|
8IF2
 
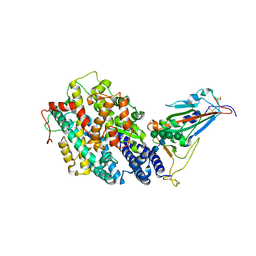 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Omicron BQ.1.1 variant spike protein in complex with its receptor ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Kimura, K, Suzuki, T, Hashiguchi, T. | | 登録日 | 2023-02-17 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-05-24 | | 実験手法 | X-RAY DIFFRACTION (2.78 Å) | | 主引用文献 | Convergent evolution of SARS-CoV-2 Omicron subvariants leading to the emergence of BQ.1.1 variant.
Nat Commun, 14, 2023
|
|
8IOV
 
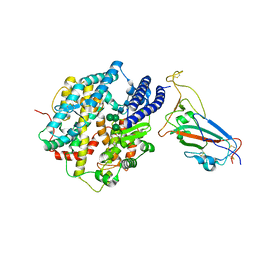 | | Structure of SARS-CoV-2 XBB.1 spike RBD in complex with ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Anraku, Y, Kita, S, Yajima, H, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | 登録日 | 2023-03-13 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (3.29 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants.
Nat Commun, 14, 2023
|
|
1GU5
 
 | |
1GTW
 
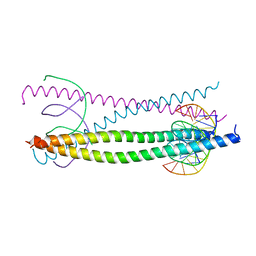 | |
1GU4
 
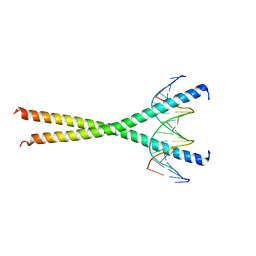 | |
1H89
 
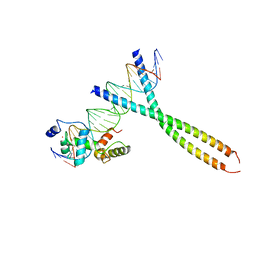 | | CRYSTAL STRUCTURE OF TERNARY PROTEIN-DNA COMPLEX2 | | 分子名称: | CAAT/ENHANCER BINDING PROTEIN BETA, DNA(5'-(*CP*CP*AP*GP*TP*CP*CP*GP*TP*TP*AP* AP*GP*GP*AP*TP*TP*GP*CP*GP*CP*CP*AP*CP*AP*T)-3'), DNA(5'-(*GP*AP*TP*GP*TP*GP*GP*CP*GP*CP*AP* AP*TP*CP*CP*TP*TP*AP*AP*CP*GP*GP*AP*CP*TP*G)-3'), ... | | 著者 | Tahirov, T.H, Ogata, K. | | 登録日 | 2001-01-30 | | 公開日 | 2002-01-28 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Mechanism of C-Myb-C/Ebpbeta Cooperation from Separated Sites on a Promoter
Cell(Cambridge,Mass.), 108, 2002
|
|
1H8A
 
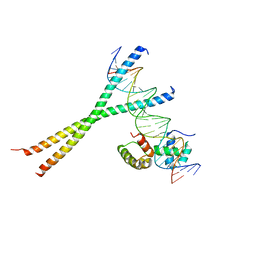 | | CRYSTAL STRUCTURE OF TERNARY PROTEIN-DNA COMPLEX3 | | 分子名称: | CAAT/ENHANCER BINDING PROTEIN BETA, DNA(5'-(*CP*CP*AP*GP*TP*CP*CP*GP*TP*TP*AP* AP*GP*GP*AP*TP*TP*GP*CP*GP*CP*CP*AP*CP*AP*T)-3'), DNA(5'-(*GP*AP*TP*GP*TP*GP*GP*CP*GP*CP*AP* AP*TP*CP*CP*TP*TP*AP*AP*CP*GP*GP*AP*CP*TP*G)-3'), ... | | 著者 | Tahirov, T.H, Ogata, K. | | 登録日 | 2001-01-31 | | 公開日 | 2002-01-28 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | Mechanism of C-Myb-C/Ebpbeta Cooperation from Separated Sites on a Promoter
Cell(Cambridge,Mass.), 108, 2002
|
|
8GV2
 
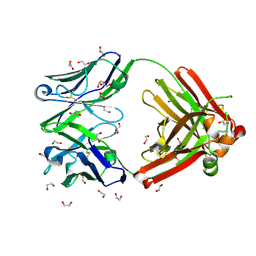 | | Crystal structure of anti-FX IgG fab without FAST-Ig mutations | | 分子名称: | 1,2-ETHANEDIOL, Anti-factor X IgG fab heavy chain, Anti-factor X IgG fab light chain | | 著者 | Koga, H, Yamano, T, Fukami, T.A, Sampei, Z, Shiraiwa, H, Torizawa, T. | | 登録日 | 2022-09-14 | | 公開日 | 2023-06-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.274 Å) | | 主引用文献 | Efficient production of bispecific antibody by FAST-Ig TM and its application to NXT007 for the treatment of hemophilia A.
Mabs, 15, 2023
|
|
8GV0
 
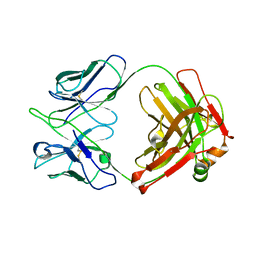 | | Crystal structure of anti-FIXa IgG fab without FAST-Ig mutations | | 分子名称: | Anti-factor IXa IgG fab heavy chain, Anti-factor IXa IgG fab light chain | | 著者 | Koga, H, Yamano, T, Fukami, T.A, Sampei, Z, Shiraiwa, H, Torizawa, T. | | 登録日 | 2022-09-14 | | 公開日 | 2023-06-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.192 Å) | | 主引用文献 | Efficient production of bispecific antibody by FAST-Ig TM and its application to NXT007 for the treatment of hemophilia A.
Mabs, 15, 2023
|
|
8GV1
 
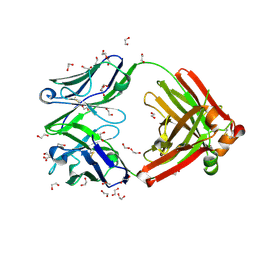 | | Crystal structure of anti-FX IgG fab with FAST-Ig mutations | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, Anti-factor X IgG fab heavy chain, ... | | 著者 | Koga, H, Yamano, T, Fukami, T.A, Sampei, Z, Shiraiwa, H, Torizawa, T. | | 登録日 | 2022-09-14 | | 公開日 | 2023-06-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.186 Å) | | 主引用文献 | Efficient production of bispecific antibody by FAST-Ig TM and its application to NXT007 for the treatment of hemophilia A.
Mabs, 15, 2023
|
|
8GUZ
 
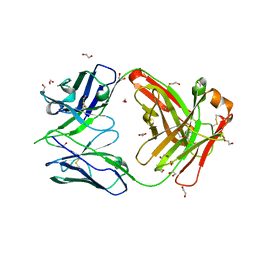 | | Crystal structure of anti-FIXa IgG fab with FAST-Ig mutations | | 分子名称: | 1,2-ETHANEDIOL, Anti-factor IXa IgG fab heavy chain, Anti-factor IXa IgG fab light chain | | 著者 | Koga, H, Yamano, T, Fukami, T.A, Sampei, Z, Shiraiwa, H, Torizawa, T. | | 登録日 | 2022-09-14 | | 公開日 | 2023-06-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Efficient production of bispecific antibody by FAST-Ig TM and its application to NXT007 for the treatment of hemophilia A.
Mabs, 15, 2023
|
|
7C03
 
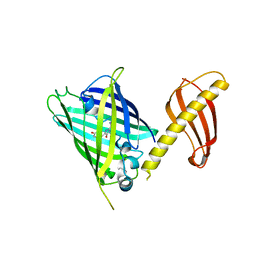 | | Crystal structure of POLArISact(T57S), genetically encoded probe for fluorescent polarization | | 分子名称: | POLArISact(T57S) | | 著者 | Tomabechi, Y, Sakai, N, Shirouzu, M. | | 登録日 | 2020-04-30 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.501 Å) | | 主引用文献 | POLArIS, a versatile probe for molecular orientation, revealed actin filaments associated with microtubule asters in early embryos.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1H88
 
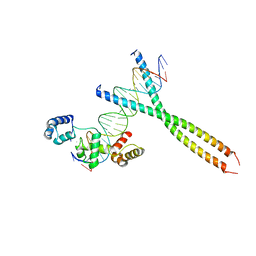 | | CRYSTAL STRUCTURE OF TERNARY PROTEIN-DNA COMPLEX1 | | 分子名称: | AMMONIUM ION, CCAAT/ENHANCER BINDING PROTEIN BETA, DNA(5'-(*CP*CP*AP*GP*TP*CP*CP*GP*TP*TP*AP* AP*GP*GP*AP*TP*TP*GP*CP*GP*CP*CP*AP*CP*AP*T)-3'), ... | | 著者 | Tahirov, T.H, Ogata, K. | | 登録日 | 2001-01-29 | | 公開日 | 2002-01-28 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Mechanism of C-Myb-C/Ebpbeta Cooperation from Separated Sites on a Promoter
Cell(Cambridge,Mass.), 108, 2002
|
|
4DGU
 
 | |
4EPS
 
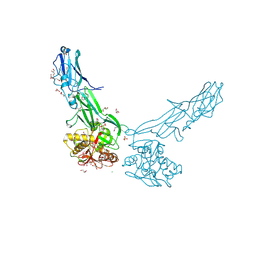 | |
6OCZ
 
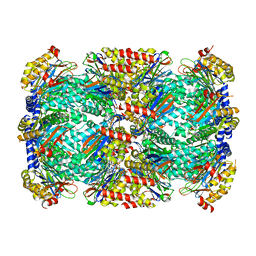 | | Crystal Structure of Mycobacterium tuberculosis Proteasome in Complex with Phenylimidazole-based Inhibitor A86 | | 分子名称: | CITRIC ACID, DIMETHYLFORMAMIDE, N-{(2S)-1-({(2S)-1-[(2,4-difluorobenzyl)amino]-1-oxopropan-2-yl}amino)-1,4-dioxo-4-[(2R)-2-phenylpyrrolidin-1-yl]butan-2-yl}-5-methyl-1,2-oxazole-3-carboxamide (non-preferred name), ... | | 著者 | Hsu, H.C, Li, H. | | 登録日 | 2019-03-25 | | 公開日 | 2019-10-09 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Selective Phenylimidazole-Based Inhibitors of theMycobacterium tuberculosisProteasome.
J.Med.Chem., 62, 2019
|
|
6OCW
 
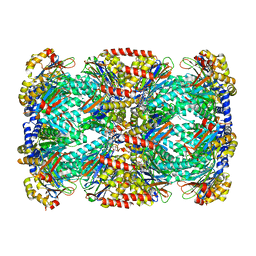 | | Crystal Structure of Mycobacterium tuberculosis Proteasome in Complex with Phenylimidazole-based Inhibitor A85 | | 分子名称: | CITRIC ACID, DIMETHYLFORMAMIDE, N-{(2S)-1-({(2S)-1-[(2,4-difluorobenzyl)amino]-1-oxopropan-2-yl}amino)-4-[(2S)-2-methylpiperidin-1-yl]-1,4-dioxobutan-2-yl}-5-methyl-1,2-oxazole-3-carboxamide (non-preferred name), ... | | 著者 | Hsu, H.C, Li, H. | | 登録日 | 2019-03-25 | | 公開日 | 2019-10-09 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Selective Phenylimidazole-Based Inhibitors of theMycobacterium tuberculosisProteasome.
J.Med.Chem., 62, 2019
|
|
3T2L
 
 | |
3SY6
 
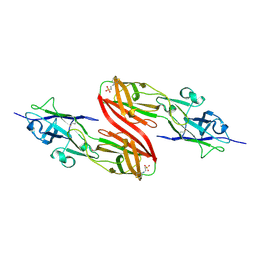 | |
3R4R
 
 | |
8GS6
 
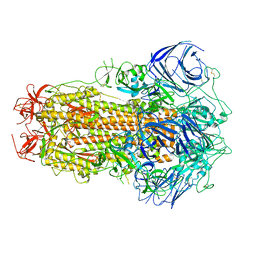 | | Structure of the SARS-CoV-2 BA.2.75 spike glycoprotein (closed state 1) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Anraku, Y, Tabata-Sasaki, K, Kita, S, Fukuhara, H, Maenaka, K, Hashiguchi, T. | | 登録日 | 2022-09-05 | | 公開日 | 2022-10-26 | | 最終更新日 | 2023-03-29 | | 実験手法 | ELECTRON MICROSCOPY (2.86 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 Omicron BA.2.75 variant.
Cell Host Microbe, 30, 2022
|
|
6ODE
 
 | |
