8CGB
 
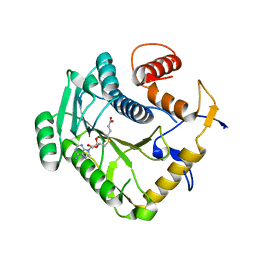 | | Monkeypox virus VP39 in complex with SAH | | 分子名称: | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Silhan, J, Klima, M, Skvara, P, Boura, E. | | 登録日 | 2023-02-03 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | Discovery and structural characterization of monkeypox virus methyltransferase VP39 inhibitors reveal similarities to SARS-CoV-2 nsp14 methyltransferase.
Nat Commun, 14, 2023
|
|
4RZD
 
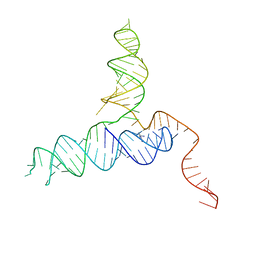 | | Crystal Structure of a PreQ1 Riboswitch | | 分子名称: | 7-DEAZA-7-AMINOMETHYL-GUANINE, PreQ1-III Riboswitch (Class 3) | | 著者 | Wedekind, J.E, Liberman, J.A, Salim, M. | | 登録日 | 2014-12-20 | | 公開日 | 2015-07-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structural analysis of a class III preQ1 riboswitch reveals an aptamer distant from a ribosome-binding site regulated by fast dynamics.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5D5L
 
 | |
3REW
 
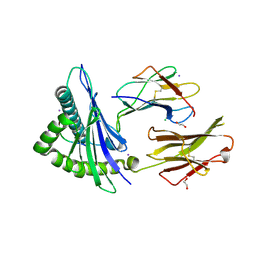 | | Crystal structure of an lmp2a-derived peptide bound to human class i mhc hla-a2 | | 分子名称: | 1,2-ETHANEDIOL, Beta-2-microglobulin, CHLORIDE ION, ... | | 著者 | Wood, A, Mohammed, F, Salim, M, Tranter, A, Rickinson, A.B, Moss, P.A.H, Stauss, H.J, Steven, N.M, Willcox, B.E. | | 登録日 | 2011-04-05 | | 公開日 | 2012-01-18 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural and energetic evidence for highly peptide-specific tumor antigen targeting via allo-MHC restriction.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3REV
 
 | | Crystal structure of human alloreactive tcr nb20 | | 分子名称: | 1,2-ETHANEDIOL, TCR NB20 ALPHA CHAIN, TCR NB20 BETA CHAIN | | 著者 | Wood, A, Mohammed, F, Salim, M, Tranter, A, Rickinson, A.B, Moss, P.A.H, Stauss, H.J, Steven, N.M, Willcox, B.E. | | 登録日 | 2011-04-05 | | 公開日 | 2012-01-18 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural and energetic evidence for highly peptide-specific tumor antigen targeting via allo-MHC restriction.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2VVI
 
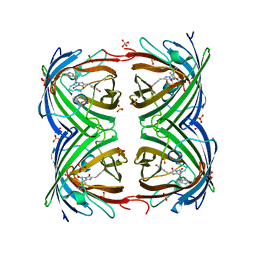 | | IrisFP fluorescent protein in its green form, trans conformation | | 分子名称: | Green to red photoconvertible GFP-like protein EosFP, SULFATE ION, SULFITE ION | | 著者 | Adam, V, Lelimousin, M, Boehme, S, Desfonds, G, Nienhaus, K, Field, M.J, Wiedenmann, J, McSweeney, S, Nienhaus, G.U, Bourgeois, D. | | 登録日 | 2008-06-09 | | 公開日 | 2008-11-11 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Characterization of Irisfp, an Optical Highlighter Undergoing Multiple Photo-Induced Transformations.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2VVH
 
 | | IrisFP fluorescent protein in its green form, cis conformation | | 分子名称: | Green to red photoconvertible GFP-like protein EosFP, SULFATE ION, SULFITE ION | | 著者 | Adam, V, Lelimousin, M, Boehme, S, Desfonds, G, Nienhaus, K, Field, M.J, Wiedenmann, J, McSweeney, S, Nienhaus, G.U, Bourgeois, D. | | 登録日 | 2008-06-09 | | 公開日 | 2008-11-11 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural Characterization of Irisfp, an Optical Highlighter Undergoing Multiple Photo-Induced Transformations.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2VVJ
 
 | | IrisFP fluorescent protein in its red form, cis conformation | | 分子名称: | Green to red photoconvertible GFP-like protein EosFP, SULFATE ION, SULFITE ION | | 著者 | Adam, V, Lelimousin, M, Boehme, S, Desfonds, G, Nienhaus, K, Field, M.J, Wiedenmann, J, McSweeney, S, Nienhaus, G.U, Bourgeois, D. | | 登録日 | 2008-06-09 | | 公開日 | 2008-08-12 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Characterization of Irisfp, an Optical Highlighter Undergoing Multiple Photo-Induced Transformations.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
1LJ8
 
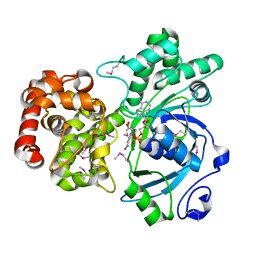 | | Crystal structure of mannitol dehydrogenase in complex with NAD | | 分子名称: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, mannitol dehydrogenase | | 著者 | Kavanagh, K.L, Klimacek, M, Nidetzky, B, Wilson, D.K. | | 登録日 | 2002-04-19 | | 公開日 | 2002-11-15 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of Pseudomonas fluorescens mannitol 2-dehydrogenase binary and ternary
complexes. Specificity and catalytic mechanism
J.Biol.Chem., 277, 2002
|
|
1JEZ
 
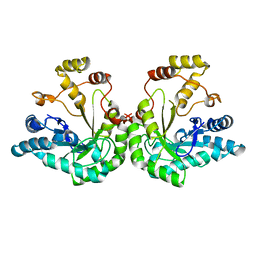 | | THE STRUCTURE OF XYLOSE REDUCTASE, A DIMERIC ALDO-KETO REDUCTASE FROM CANDIDA TENUIS | | 分子名称: | XYLOSE REDUCTASE | | 著者 | Kavanagh, K.L, Klimacek, M, Nidetzky, B, Wilson, D.K. | | 登録日 | 2001-06-19 | | 公開日 | 2002-07-05 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The structure of apo and holo forms of xylose reductase, a dimeric aldo-keto reductase from Candida tenuis.
Biochemistry, 41, 2002
|
|
1M2W
 
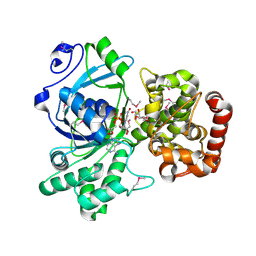 | | Pseudomonas fluorescens mannitol 2-dehydrogenase ternary complex with NAD and D-mannitol | | 分子名称: | D-MANNITOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, mannitol dehydrogenase | | 著者 | Kavanagh, K.L, Klimacek, M, Nidetzky, B, Wilson, D.K. | | 登録日 | 2002-06-25 | | 公開日 | 2002-11-15 | | 最終更新日 | 2011-11-16 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of Pseudomonas fluorescens Mannitol 2-Dehydrogenase Binary and Ternary Complexes. Specificity and Catalytic Mechanism
J.Biol.Chem., 277, 2002
|
|
1MI3
 
 | | 1.8 Angstrom structure of xylose reductase from Candida tenuis in complex with NAD | | 分子名称: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, xylose reductase | | 著者 | Kavanagh, K.L, Klimacek, M, Nidetzky, B, Wilson, D.K. | | 登録日 | 2002-08-21 | | 公開日 | 2003-08-05 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of xylose reductase bound to NAD+ and the basis for single and dual co-substrate specificity in family 2 aldo-keto reductases
Biochem.J., 373, 2003
|
|
1K8C
 
 | | Crystal structure of dimeric xylose reductase in complex with NADP(H) | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, xylose reductase | | 著者 | Kavanagh, K.L, Klimacek, M, Nidetzky, B, Wilson, D.K. | | 登録日 | 2001-10-23 | | 公開日 | 2002-07-05 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The structure of apo and holo forms of xylose reductase, a dimeric aldo-keto reductase from Candida tenuis.
Biochemistry, 41, 2002
|
|
2N73
 
 | | Solution structure of the ACBD3:PI4KB complex | | 分子名称: | Golgi resident protein GCP60, Phosphatidylinositol 4-kinase beta | | 著者 | Veverka, V, Hexnerova, R. | | 登録日 | 2015-09-02 | | 公開日 | 2016-07-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural insights and in vitro reconstitution of membrane targeting and activation of human PI4KB by the ACBD3 protein.
Sci Rep, 6, 2016
|
|
2N72
 
 | |
4UZV
 
 | | Structure of a triple mutant of ASV-TfTrHb | | 分子名称: | ACETATE ION, HEMOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Baiocco, P, Bonamore, A, Sciamanna, N, Ilari, A, Boechi, L, Boffi, A, Smulevich, G, Feis, A. | | 登録日 | 2014-09-09 | | 公開日 | 2014-09-17 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Role of Local Structure and Dynamics of Small Ligand Migration in Proteins: A Study of a Mutated Truncated Hemoprotein from Thermobifida Fusca by Time Resolved Mir Spectroscopy.
J.Phys.Chem.B, 118, 2014
|
|
4PLA
 
 | |
3MLE
 
 | | Crystal structure of dethiobiotin synthetase (BioD) from Helicobacter pylori cocrystallized with ATP | | 分子名称: | 8-aminooctanoic acid, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | 著者 | Nicholls, R, Porebski, P.J, Klimecka, M.M, Chruszcz, M, Murzyn, K, Joachimiak, A, Murshudov, G, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-04-16 | | 公開日 | 2010-05-19 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals differences between family members.
Febs J., 279, 2012
|
|
3QXS
 
 | | Crystal structure of dethiobiotin synthetase (BioD) from Helicobacter pylori complexed with ANP | | 分子名称: | 1,2-ETHANEDIOL, Dethiobiotin synthetase, MAGNESIUM ION, ... | | 著者 | Klimecka, M.M, Porebski, P.J, Chruszcz, M, Jablonska, K, Murzyn, K, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-03-02 | | 公開日 | 2011-03-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals differences between family members.
Febs J., 279, 2012
|
|
3QXC
 
 | | Crystal structure of dethiobiotin synthetase (BioD) from Helicobacter pylori complexed with ATP | | 分子名称: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Porebski, P.J, Klimecka, M.M, Chruszcz, M, Murzyn, K, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-03-01 | | 公開日 | 2011-03-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.34 Å) | | 主引用文献 | Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals differences between family members.
Febs J., 279, 2012
|
|
3QXJ
 
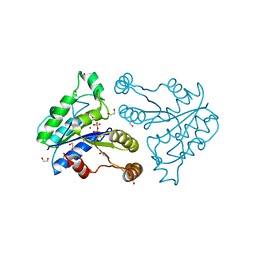 | | Crystal structure of dethiobiotin synthetase (BioD) from Helicobacter pylori complexed with GTP | | 分子名称: | 1,2-ETHANEDIOL, Dethiobiotin synthetase, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Klimecka, M.M, Porebski, P.J, Chruszcz, M, Murzyn, K, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-03-01 | | 公開日 | 2011-03-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals differences between family members.
Febs J., 279, 2012
|
|
3QY0
 
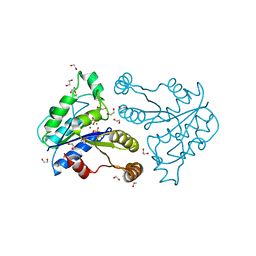 | | Crystal structure of dethiobiotin synthetase (BioD) from Helicobacter pylori complexed with GDP | | 分子名称: | 1,2-ETHANEDIOL, Dethiobiotin synthetase, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Porebski, P.J, Klimecka, M.M, Chruszcz, M, Murzyn, K, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-03-02 | | 公開日 | 2011-03-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals differences between family members.
Febs J., 279, 2012
|
|
5KOR
 
 | | Arabidopsis thaliana fucosyltransferase 1 (FUT1) in complex with GDP and a xylo-oligossacharide | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Rocha, J, de Sanctis, D, Breton, C. | | 登録日 | 2016-07-01 | | 公開日 | 2016-10-12 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of Arabidopsis thaliana FUT1 Reveals a Variant of the GT-B Class Fold and Provides Insight into Xyloglucan Fucosylation.
Plant Cell, 28, 2016
|
|
4YC9
 
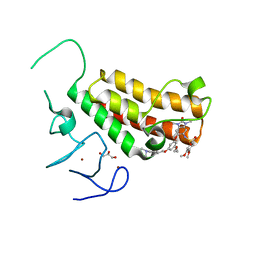 | | Crystal structure of TRIM24 PHD-bromodomain complexed with N-(6-{3-[4-(dimethylamino)butoxy]-5-propoxyphenoxy}-1,3-dimethyl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl)-3,4-dimethoxybenzene-1-sulfonamide (8i) | | 分子名称: | GLYCEROL, N-(6-{3-[4-(dimethylamino)butoxy]-5-propoxyphenoxy}-1,3-dimethyl-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl)-3,4-dimethoxybenzenesulfonamide, Transcription intermediary factor 1-alpha, ... | | 著者 | Poncet-Montange, G, Palmer, W, Jones, P. | | 登録日 | 2015-02-19 | | 公開日 | 2015-06-24 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Structure-Guided Design of IACS-9571, a Selective High-Affinity Dual TRIM24-BRPF1 Bromodomain Inhibitor.
J.Med.Chem., 59, 2016
|
|
4YAB
 
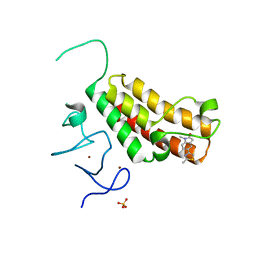 | | Crystal structure of TRIM24 PHD-bromodomain complexed with 1-methyl-5-(2-methyl-1 3-thiazol-4-yl)-2 3-dihydro-1H-indol-2-one (1) | | 分子名称: | 1-methyl-5-(2-methyl-1,3-thiazol-4-yl)-1,3-dihydro-2H-indol-2-one, SULFATE ION, Transcription intermediary factor 1-alpha, ... | | 著者 | Poncet-Montange, G, Palmer, W, Jones, P. | | 登録日 | 2015-02-17 | | 公開日 | 2015-06-24 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure-Guided Design of IACS-9571, a Selective High-Affinity Dual TRIM24-BRPF1 Bromodomain Inhibitor.
J.Med.Chem., 59, 2016
|
|
