5MPP
 
 | | Structure of AaLS-wt | | 分子名称: | 6,7-dimethyl-8-ribityllumazine synthase | | 著者 | Sasaki, E, Boehringer, D, Leibundgut, M, Ban, N, Hilvert, D. | | 登録日 | 2016-12-17 | | 公開日 | 2017-03-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structure and assembly of scalable porous protein cages.
Nat Commun, 8, 2017
|
|
5MQ3
 
 | | Structure of AaLS-neg | | 分子名称: | 6,7-dimethyl-8-ribityllumazine synthase | | 著者 | Sasaki, E, Boehringer, D, Leibundgut, M, Ban, N, Hilvert, D. | | 登録日 | 2016-12-20 | | 公開日 | 2017-03-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (5.4 Å) | | 主引用文献 | Structure and assembly of scalable porous protein cages.
Nat Commun, 8, 2017
|
|
5MQ7
 
 | | Structure of AaLS-13 | | 分子名称: | 6,7-dimethyl-8-ribityllumazine synthase | | 著者 | Sasaki, E, Boehringer, D, Leibundgut, M, Ban, N, Hilvert, D. | | 登録日 | 2016-12-20 | | 公開日 | 2017-03-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (5.2 Å) | | 主引用文献 | Structure and assembly of scalable porous protein cages.
Nat Commun, 8, 2017
|
|
5NCO
 
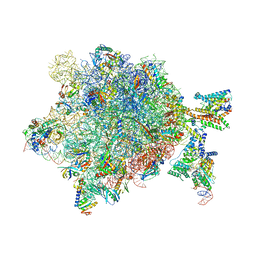 | | Quaternary complex between SRP, SR, and SecYEG bound to the translating ribosome | | 分子名称: | 23S rRNA, 4.5S SRP RNA (Ffs), 50S ribosomal protein L10, ... | | 著者 | Jomaa, A, Hwang Fu, Y, Boerhinger, D, Leibundgut, M, Shan, S.O, Ban, N. | | 登録日 | 2017-03-06 | | 公開日 | 2017-05-24 | | 最終更新日 | 2018-03-28 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Structure of the quaternary complex between SRP, SR, and translocon bound to the translating ribosome.
Nat Commun, 8, 2017
|
|
5O61
 
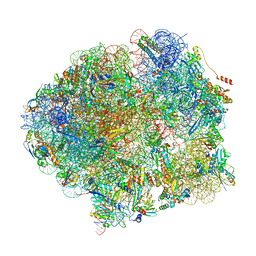 | | The complete structure of the Mycobacterium smegmatis 70S ribosome | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Hentschel, J, Burnside, C, Mignot, I, Leibundgut, M, Boehringer, D, Ban, N. | | 登録日 | 2017-06-03 | | 公開日 | 2017-07-12 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.31 Å) | | 主引用文献 | The Complete Structure of the Mycobacterium smegmatis 70S Ribosome.
Cell Rep, 20, 2017
|
|
6YXX
 
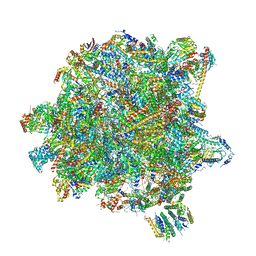 | | State A of the Trypanosoma brucei mitoribosomal large subunit assembly intermediate | | 分子名称: | 12S ribosomal RNA, 50S ribosomal protein L13, putative, ... | | 著者 | Jaskolowski, M, Ramrath, D.J.F, Bieri, P, Niemann, M, Mattei, S, Calderaro, S, Leibundgut, M.A, Horn, E.K, Boehringer, D, Schneider, A, Ban, N. | | 登録日 | 2020-05-04 | | 公開日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural Insights into the Mechanism of Mitoribosomal Large Subunit Biogenesis.
Mol.Cell, 79, 2020
|
|
6YXY
 
 | | State B of the Trypanosoma brucei mitoribosomal large subunit assembly intermediate | | 分子名称: | 12S ribosomal RNA, ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Jaskolowski, M, Ramrath, D.J.F, Bieri, P, Niemann, M, Mattei, S, Calderaro, S, Leibundgut, M.A, Horn, E.K, Boehringer, D, Schneider, A, Ban, N. | | 登録日 | 2020-05-04 | | 公開日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural Insights into the Mechanism of Mitoribosomal Large Subunit Biogenesis.
Mol.Cell, 79, 2020
|
|
5A2Q
 
 | | Structure of the HCV IRES bound to the human ribosome | | 分子名称: | 18S RRNA, HCV IRES, MAGNESIUM ION, ... | | 著者 | Quade, N, Leiundgut, M, Boehringer, D, Heuvel, J.v.d, Ban, N. | | 登録日 | 2015-05-21 | | 公開日 | 2015-07-15 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Cryo-Em Structure of Hepatitis C Virus Ires Bound to the Human Ribosome at 3.9 Angstrom Resolution
Nat.Commun., 6, 2015
|
|
6ZOL
 
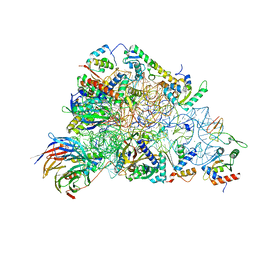 | | SARS-CoV-2-Nsp1-40S complex, focused on head | | 分子名称: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S12, ... | | 著者 | Schubert, K, Karousis, E.D, Jomaa, A, Scaiola, A, Echeverria, B, Gurzeler, L.-A, Leibundgut, M.L, Thiel, V, Muehlemann, O, Ban, N. | | 登録日 | 2020-07-07 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZOJ
 
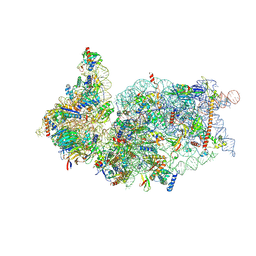 | | SARS-CoV-2-Nsp1-40S complex, composite map | | 分子名称: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | 著者 | Schubert, K, Karousis, E.D, Jomaa, A, Scaiola, A, Echeverria, B, Gurzeler, L.-A, Leibundgut, M.L, Thiel, V, Muehlemann, O, Ban, N. | | 登録日 | 2020-07-07 | | 公開日 | 2020-07-22 | | 最終更新日 | 2021-02-10 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7A4F
 
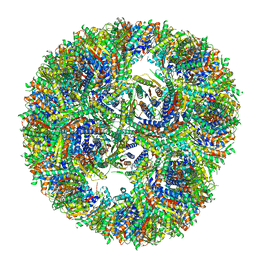 | |
7A4H
 
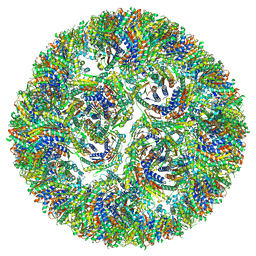 | |
7A4G
 
 | |
7A4I
 
 | |
7A4J
 
 | |
7O7Y
 
 | | Rabbit 80S ribosome stalled close to the mutated SARS-CoV-2 slippery site by a pseudoknot (high resolution) | | 分子名称: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | 著者 | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | 登録日 | 2021-04-14 | | 公開日 | 2021-06-02 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.2 Å) | | 主引用文献 | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
7O80
 
 | | Rabbit 80S ribosome in complex with eRF1 and ABCE1 stalled at the STOP codon in the mutated SARS-CoV-2 slippery site | | 分子名称: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | 著者 | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | 登録日 | 2021-04-14 | | 公開日 | 2021-06-02 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
7O81
 
 | | Rabbit 80S ribosome colliding in another ribosome stalled by the SARS-CoV-2 pseudoknot | | 分子名称: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | 著者 | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | 登録日 | 2021-04-14 | | 公開日 | 2021-06-02 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
7O7Z
 
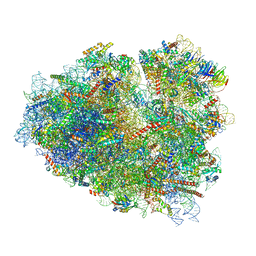 | | Rabbit 80S ribosome stalled close to the mutated SARS-CoV-2 slippery site by a pseudoknot (classified for pseudoknot) | | 分子名称: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | 著者 | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | 登録日 | 2021-04-14 | | 公開日 | 2021-06-02 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.4 Å) | | 主引用文献 | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
2ML8
 
 | |
7OYH
 
 | |
7OYF
 
 | |
7OY3
 
 | |
7OXY
 
 | | Crystal structure of depupylase Dop in complex with Pup and AMP-PCP | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Cui, H. | | 登録日 | 2021-06-23 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structures of prokaryotic ubiquitin-like protein Pup in complex with depupylase Dop reveal the mechanism of catalytic phosphate formation.
Nat Commun, 12, 2021
|
|
7OXV
 
 | | Crystal structure of depupylase Dop in the Dop-loop-inserted state | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Depupylase, ... | | 著者 | Cui, H. | | 登録日 | 2021-06-23 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.394 Å) | | 主引用文献 | Structures of prokaryotic ubiquitin-like protein Pup in complex with depupylase Dop reveal the mechanism of catalytic phosphate formation.
Nat Commun, 12, 2021
|
|
