5HC4
 
 | | Structure of esterase Est22 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Lipolytic enzyme | | 著者 | Li, J, Huang, J. | | 登録日 | 2016-01-04 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural insights of a hormone sensitive lipase homologue Est22.
Sci Rep, 6, 2016
|
|
5HC0
 
 | |
5HC5
 
 | |
5HC3
 
 | | The structure of esterase Est22 | | 分子名称: | GLYCEROL, Lipolytic enzyme | | 著者 | Li, J, Huang, J. | | 登録日 | 2016-01-04 | | 公開日 | 2017-01-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural insights of a hormone sensitive lipase homologue Est22.
Sci Rep, 6, 2016
|
|
5HC2
 
 | |
8THH
 
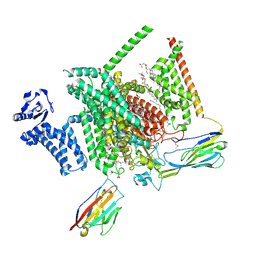 | | Cryo-EM structure of Nav1.7 with LTG | | 分子名称: | (6M)-6-(2,3-dichlorophenyl)-1,2,4-triazine-3,5-diamine, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | 著者 | Fan, X, Huang, J, Yan, N. | | 登録日 | 2023-07-16 | | 公開日 | 2023-11-22 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Dual-pocket inhibition of Na v channels by the antiepileptic drug lamotrigine.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8THG
 
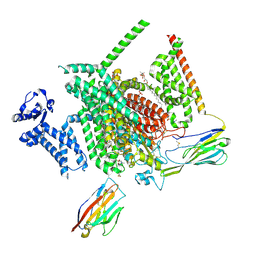 | | Cryo-EM structure of Nav1.7 with RLZ | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Fan, X, Huang, J, Yan, N. | | 登録日 | 2023-07-16 | | 公開日 | 2023-11-22 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Dual-pocket inhibition of Na v channels by the antiepileptic drug lamotrigine.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5C13
 
 | |
4I80
 
 | |
5GP4
 
 | |
5H2Z
 
 | |
4G6F
 
 | | Crystal Structure of 10E8 Fab in Complex with an HIV-1 gp41 Peptide | | 分子名称: | 10E8 Heavy Chain, 10E8 Light Chain, gp41 MPER Peptide | | 著者 | Ofek, G, Huang, J, Connors, M, Kwong, P.D. | | 登録日 | 2012-07-19 | | 公開日 | 2012-09-26 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Broad and potent neutralization of HIV-1 by a gp41-specific human antibody.
Nature, 491, 2012
|
|
6LT7
 
 | |
6NTF
 
 | | Crystal structure of a computationally optimized H5 influenza hemagglutinin | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Huang, J, Bar-Peled, Y, Mousa, J.J. | | 登録日 | 2019-01-29 | | 公開日 | 2019-09-18 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural and antigenic characterization of a computationally-optimized H5 hemagglutinin influenza vaccine.
Vaccine, 37, 2019
|
|
6KIX
 
 | | Cryo-EM structure of human MLL1-NCP complex, binding mode1 | | 分子名称: | DNA (145-MER), GLUTAMINE, Histone H2A, ... | | 著者 | Huang, J, Xue, H, Yao, T. | | 登録日 | 2019-07-20 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
6KIV
 
 | | Cryo-EM structure of human MLL1-ubNCP complex (4.0 angstrom) | | 分子名称: | DNA (145-MER), Histone H2A, Histone H2B 1.1, ... | | 著者 | Huang, J, Xue, H, Yao, T. | | 登録日 | 2019-07-20 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
6KIU
 
 | | Cryo-EM structure of human MLL1-ubNCP complex (3.2 angstrom) | | 分子名称: | DNA (145-MER), GLUTAMINE, Histone H2A, ... | | 著者 | Huang, J, Xue, H, Yao, T. | | 登録日 | 2019-07-20 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
6KIZ
 
 | | Cryo-EM structure of human MLL1-NCP complex, binding mode2 | | 分子名称: | DNA (145-MER), Histone H2A, Histone H2B 1.1, ... | | 著者 | Huang, J, Xue, H, Yao, T. | | 登録日 | 2019-07-20 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
6KIW
 
 | | Cryo-EM structure of human MLL3-ubNCP complex (4.0 angstrom) | | 分子名称: | DNA (144-MER), DNA (145-MER), Histone H2A, ... | | 著者 | Huang, J, Xue, H, Yao, T. | | 登録日 | 2019-07-20 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
7VOH
 
 | |
4U24
 
 | | Crystal structure of the E. coli ribosome bound to dalfopristin. | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Noeske, J, Huang, J, Olivier, N.B, Giacobbe, R.A, Zambrowski, M, Cate, J.H.D. | | 登録日 | 2014-07-16 | | 公開日 | 2014-07-30 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Synergy of streptogramin antibiotics occurs independently of their effects on translation.
Antimicrob.Agents Chemother., 58, 2014
|
|
4U1U
 
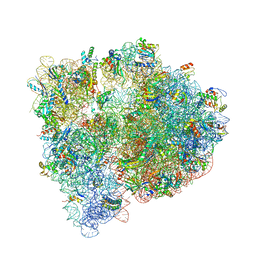 | | Crystal structure of the E. coli ribosome bound to quinupristin. | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Noeske, J, Huang, J, Olivier, N.B, Giacobbe, R.A, Zambrowski, M, Cate, J.H.D. | | 登録日 | 2014-07-16 | | 公開日 | 2014-07-30 | | 最終更新日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Synergy of streptogramin antibiotics occurs independently of their effects on translation.
Antimicrob.Agents Chemother., 58, 2014
|
|
4UX5
 
 | | Structure of DNA complex of PCG2 | | 分子名称: | 5'-D(*CP*AP*AP*TP*GP*AP*CP*GP*CP*GP*TP*AP*AP*GP)-3', 5'-D(*CP*TP*TP*AP*CP*GP*CP*GP*TP*CP*AP*TP*TP*GP)-3', TRANSCRIPTION FACTOR MBP1 | | 著者 | Liu, J, Huang, J, Zhao, Y, Liu, H, Wang, D, Yang, J, Zhao, W, Taylor, I.A, Peng, Y. | | 登録日 | 2014-08-19 | | 公開日 | 2015-01-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural Basis of DNA Recognition by Pcg2 Reveals a Novel DNA Binding Mode for Winged Helix-Turn-Helix Domains.
Nucleic Acids Res., 43, 2015
|
|
4KN8
 
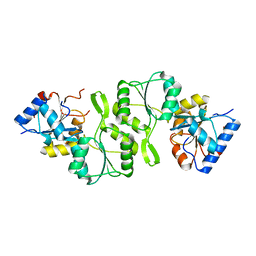 | | Crystal structure of Bs-TpNPPase | | 分子名称: | Thermostable NPPase | | 著者 | Guo, Z, Wang, F, Huang, J, Gong, W, Ji, C. | | 登録日 | 2013-05-09 | | 公開日 | 2014-04-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.502 Å) | | 主引用文献 | Crystal Structure of Thermostable p-nitrophenylphosphatase from Bacillus Stearothermophilus (Bs-TpNPPase)
PROTEIN PEPT.LETT., 21, 2014
|
|
4U26
 
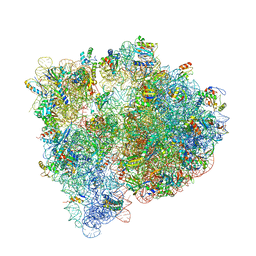 | | Crystal structure of the E. coli ribosome bound to dalfopristin and quinupristin. | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Noeske, J, Huang, J, Olivier, N.B, Giacobbe, R.A, Zambrowski, M, Cate, J.H.D. | | 登録日 | 2014-07-16 | | 公開日 | 2014-07-30 | | 最終更新日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Synergy of streptogramin antibiotics occurs independently of their effects on translation.
Antimicrob.Agents Chemother., 58, 2014
|
|
