4XA3
 
 | | Crystal structure of the coiled-coil surrounding Skip 2 of MYH7 | | 分子名称: | Gp7-MYH7(1361-1425)-Eb1 chimera protein | | 著者 | Taylor, K.C, Buvoli, M, Korkmaz, E.N, Buvoli, A, Zheng, Y, Heinz, N.T, Qiang, C, Leinwand, L.A, Rayment, I. | | 登録日 | 2014-12-12 | | 公開日 | 2015-07-01 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.548 Å) | | 主引用文献 | Skip residues modulate the structural properties of the myosin rod and guide thick filament assembly.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6PZV
 
 | |
7L4C
 
 | | Crystal structure of the DRM2-CTT DNA complex | | 分子名称: | DNA (5'-D(*AP*TP*TP*AP*TP*TP*AP*AP*TP*(C49)P*TP*TP*AP*AP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*TP*TP*AP*AP*GP*AP*TP*TP*AP*AP*TP*AP*AP*T)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | 著者 | Fang, J, Song, J. | | 登録日 | 2020-12-18 | | 公開日 | 2021-08-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
7L4K
 
 | | Crystal structure of the DRM2-CCG DNA complex | | 分子名称: | DNA (5'-D(*AP*TP*TP*CP*CP*TP*AP*AP*TP*(C49)P*CP*GP*AP*AP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*TP*TP*CP*GP*GP*AP*TP*TP*AP*GP*GP*AP*AP*T)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | 著者 | Fang, J, Song, J. | | 登録日 | 2020-12-19 | | 公開日 | 2021-08-04 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
7L4H
 
 | | Crystal structure of the DRM2-CTG DNA complex | | 分子名称: | DNA (5'-D(*AP*TP*TP*CP*CP*TP*AP*AP*TP*(C49)P*TP*GP*AP*AP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*TP*TP*CP*AP*GP*AP*TP*TP*AP*GP*GP*AP*AP*T)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | 著者 | Fang, J, Song, J. | | 登録日 | 2020-12-19 | | 公開日 | 2021-08-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
7L4N
 
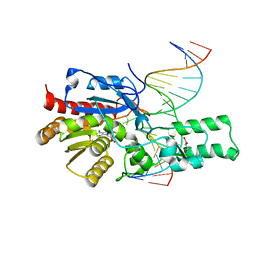 | | Crystal structure of the DRM2 (C397R)-CCG DNA complex | | 分子名称: | DNA (5'-D(*AP*TP*TP*CP*CP*TP*AP*AP*TP*(C49)P*CP*GP*AP*AP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*TP*TP*CP*GP*GP*AP*TP*TP*AP*GP*GP*AP*AP*T)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | 著者 | Fang, J, Song, J. | | 登録日 | 2020-12-19 | | 公開日 | 2021-08-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.247 Å) | | 主引用文献 | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
7L4M
 
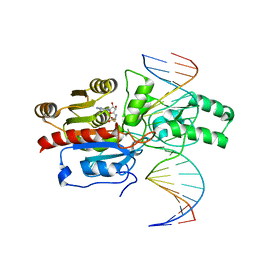 | | Crystal structure of the DRM2-CCT DNA complex | | 分子名称: | DNA (5'-D(*TP*AP*AP*AP*GP*GP*AP*GP*GP*AP*GP*GP*AP*GP*GP*AP*AP*T)-3'), DNA (5'-D(P*AP*TP*TP*CP*CP*TP*CP*CP*TP*(C49)P*CP*TP*CP*CP*TP*TP*TP*A)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | 著者 | Fang, J, Song, J. | | 登録日 | 2020-12-19 | | 公開日 | 2021-08-04 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.805 Å) | | 主引用文献 | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
7L4F
 
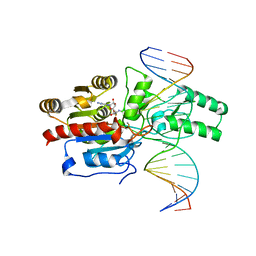 | | Crystal structure of the DRM2-CAT DNA complex | | 分子名称: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, DNA (5'-D(*AP*TP*TP*CP*CP*TP*CP*CP*TP*(C49)P*AP*TP*CP*CP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*GP*GP*AP*TP*GP*AP*GP*GP*AP*GP*GP*AP*AP*T)-3'), ... | | 著者 | Fang, J, Song, J. | | 登録日 | 2020-12-19 | | 公開日 | 2021-08-04 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
7LMM
 
 | |
7LMK
 
 | |
8U1X
 
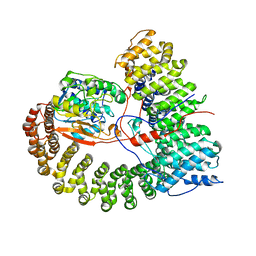 | | The structure of the PP2A-B56Delta holoenzyme mutant - E197K | | 分子名称: | MANGANESE (II) ION, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit delta isoform, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, ... | | 著者 | Wu, C.G, Xing, Y. | | 登録日 | 2023-09-04 | | 公開日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | B56 delta long-disordered arms form a dynamic PP2A regulation interface coupled with global allostery and Jordan's syndrome mutations.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
4HP1
 
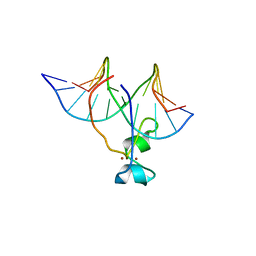 | | Crystal structure of Tet3 in complex with a non-CpG dsDNA | | 分子名称: | DNA (5'-D(*GP*CP*CP*AP*CP*(5CM)P*GP*GP*TP*GP*GP*C)-3'), LOC100036628 protein, ZINC ION | | 著者 | Chao, X, Tempel, W, Bian, C, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2012-10-23 | | 公開日 | 2012-12-05 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Tet3 CXXC Domain and Dioxygenase Activity Cooperatively Regulate Key Genes for Xenopus Eye and Neural Development.
Cell(Cambridge,Mass.), 151, 2012
|
|
4HP3
 
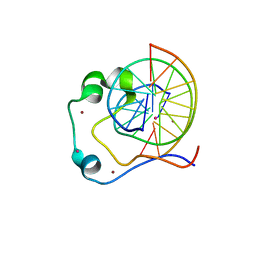 | | Crystal structure of Tet3 in complex with a CpG dsDNA | | 分子名称: | DNA (5'-D(*GP*CP*CP*AP*AP*CP*GP*TP*TP*GP*GP*C)-3'), LOC100036628 protein, UNKNOWN ATOM OR ION, ... | | 著者 | Chao, X, Tempel, W, Bian, C, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2012-10-23 | | 公開日 | 2012-12-05 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Tet3 CXXC Domain and Dioxygenase Activity Cooperatively Regulate Key Genes for Xenopus Eye and Neural Development.
Cell(Cambridge,Mass.), 151, 2012
|
|
3G2M
 
 | |
3HBN
 
 | | Crystal structure PseG-UDP complex from Campylobacter jejuni | | 分子名称: | CHLORIDE ION, GLYCEROL, UDP-sugar hydrolase, ... | | 著者 | Rangarajan, E.S, Proteau, A, Cygler, M, Matte, A, Sulea, T, Schoenhofen, I.C. | | 登録日 | 2009-05-04 | | 公開日 | 2009-05-26 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural and functional analysis of Campylobacter jejuni PseG: a udp-sugar hydrolase from the pseudaminic acid biosynthetic pathway.
J.Biol.Chem., 284, 2009
|
|
3HBM
 
 | | Crystal Structure of PseG from Campylobacter jejuni | | 分子名称: | SULFATE ION, UDP-sugar hydrolase | | 著者 | Rangarajan, E.S, Proteau, A, Cygler, M, Matte, A, Sulea, T, Schoenhofen, I.C. | | 登録日 | 2009-05-04 | | 公開日 | 2009-05-26 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural and functional analysis of Campylobacter jejuni PseG: a udp-sugar hydrolase from the pseudaminic acid biosynthetic pathway.
J.Biol.Chem., 284, 2009
|
|
3G2O
 
 | | Crystal Structure of the Glycopeptide N-methyltransferase MtfA complexed with (S)-adenosyl-L-methionine (SAM) | | 分子名称: | PCZA361.24, S-ADENOSYLMETHIONINE | | 著者 | Shi, R, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | 登録日 | 2009-01-31 | | 公開日 | 2009-05-05 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure and function of the glycopeptide N-methyltransferase MtfA, a tool for the biosynthesis of modified glycopeptide antibiotics.
Chem.Biol., 16, 2009
|
|
3E7J
 
 | |
2MMP
 
 | | Solution structure of a ribosomal protein | | 分子名称: | Uncharacterized protein | | 著者 | Feng, Y. | | 登録日 | 2014-03-17 | | 公開日 | 2014-06-11 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure determination of archaea-specific ribosomal protein L46a reveals a novel protein fold.
Biochem.Biophys.Res.Commun., 450, 2014
|
|
3CQH
 
 | |
3CQK
 
 | | Crystal Structure of L-xylulose-5-phosphate 3-epimerase UlaE (form B) complex with Zn2+ and sulfate | | 分子名称: | L-ribulose-5-phosphate 3-epimerase ulaE, SULFATE ION, ZINC ION | | 著者 | Shi, R, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | 登録日 | 2008-04-03 | | 公開日 | 2008-11-25 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | Structure of L-xylulose-5-Phosphate 3-epimerase (UlaE) from the anaerobic L-ascorbate utilization pathway of Escherichia coli: identification of a novel phosphate binding motif within a TIM barrel fold.
J.Bacteriol., 190, 2008
|
|
3CQJ
 
 | |
3CQI
 
 | |
3E80
 
 | |
3G2Q
 
 | | Crystal Structure of the Glycopeptide N-methyltransferase MtfA complexed with sinefungin | | 分子名称: | PCZA361.24, SINEFUNGIN | | 著者 | Shi, R, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | 登録日 | 2009-01-31 | | 公開日 | 2009-05-05 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Structure and function of the glycopeptide N-methyltransferase MtfA, a tool for the biosynthesis of modified glycopeptide antibiotics.
Chem.Biol., 16, 2009
|
|
