2GRB
 
 | |
8C3K
 
 | |
7A9B
 
 | | Crystal structure of Shank1 PDZ domain with ARAP3-derived peptide | | 分子名称: | 1,2-ETHANEDIOL, SH3 and multiple ankyrin repeat domains protein 1,Arf-GAP with Rho-GAP domain, ANK repeat and PH domain-containing protein 3 | | 著者 | Mariam McAuley, M, Ali, M, Ivarsson, Y, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | 登録日 | 2020-09-01 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of Shank1 PDZ domain with ARAP3-derived peptide
to be published
|
|
5LYM
 
 | |
5MM8
 
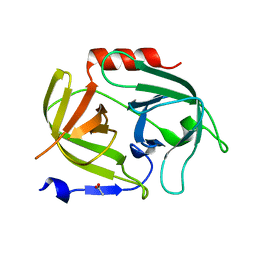 | |
5L6S
 
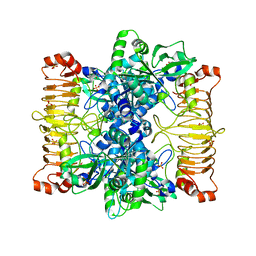 | | Crystal structure of E. coli ADP-glucose pyrophosphorylase (AGPase) in complex with a positive allosteric regulator beta-fructose-1,6-diphosphate (FBP) - AGPase*FBP | | 分子名称: | 1,6-di-O-phosphono-beta-D-fructofuranose, Glucose-1-phosphate adenylyltransferase, SULFATE ION | | 著者 | Cifuente, J.O, Albesa-Jove, D, Comino, N, Madariaga-Marcos, J, Agirre, J, Lopez-Fernandez, S, Garcia-Alija, M, Guerin, M.E. | | 登録日 | 2016-05-31 | | 公開日 | 2016-09-07 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.04 Å) | | 主引用文献 | Structural Basis of Glycogen Biosynthesis Regulation in Bacteria.
Structure, 24, 2016
|
|
5L6V
 
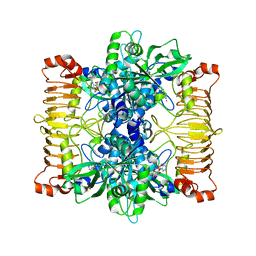 | | Crystal structure of E. coli ADP-glucose pyrophosphorylase (AGPase) in complex with a negative allosteric regulator adenosine monophosphate (AMP) - AGPase*AMP | | 分子名称: | ADENOSINE MONOPHOSPHATE, Glucose-1-phosphate adenylyltransferase, PHOSPHATE ION, ... | | 著者 | Cifuente, J.O, Albesa-Jove, D, Comino, N, Madariaga-Marcos, J, Agirre, J, Lopez-Fernandez, S, Garcia-Alija, M, Guerin, M.E. | | 登録日 | 2016-05-31 | | 公開日 | 2016-09-07 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.667 Å) | | 主引用文献 | Structural Basis of Glycogen Biosynthesis Regulation in Bacteria.
Structure, 24, 2016
|
|
5AD3
 
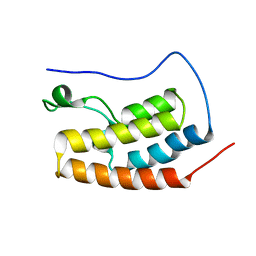 | | Bivalent binding to BET bromodomains | | 分子名称: | 3-methoxy-N-[2-[4-[1-(3-methoxy-[1,2,4]triazolo[4,3-b]pyridazin-6-yl)-4-piperidyl]phenoxy]ethyl]-N-methyl-[1,2,4]triazolo[4,3-b]pyridazin-6-amine, BROMODOMAIN-CONTAINING PROTEIN 4 | | 著者 | Waring, M.J, Chen, H, Rabow, A.A, Walker, G, Bobby, R, Boiko, S, Bradbury, R.H, Callis, R, Dale, I, Daniels, D, Flavell, L, Holdgate, G, Jowitt, T.A, Kikhney, A, McAlister, M, Ogg, D, Patel, J, Petteruti, P, Robb, G.R, Robers, M, Stratton, N, Svergun, D.I, Wang, W, Whittaker, D. | | 登録日 | 2015-08-19 | | 公開日 | 2016-09-28 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Potent and Selective Bivalent Inhibitors of Bet Bromodomains
Nat.Chem.Biol., 12, 2016
|
|
5AD2
 
 | | Bivalent binding to BET bromodomains | | 分子名称: | (3R)-4-(2-{4-[1-(3-chloro[1,2,4]triazolo[4,3-b]pyridazin-6-yl)-4-piperidinyl]phenoxy}ethyl)-1,3-dimethyl-2-piperazinone, BROMODOMAIN-CONTAINING PROTEIN 4 | | 著者 | Waring, M.J, Chen, H, Rabow, A.A, Walker, G, Bobby, R, Boiko, S, Bradbury, R.H, Callis, R, Dale, I, Daniels, D, Flavell, L, Holdgate, G, Jowitt, T.A, Kikhney, A, McAlister, M, Ogg, D, Patel, J, Petteruti, P, Robb, G.R, Robers, M, Stratton, N, Svergun, D.I, Wang, W, Whittaker, D. | | 登録日 | 2015-08-19 | | 公開日 | 2016-09-28 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Potent and Selective Bivalent Inhibitors of Bet Bromodomains
Nat.Chem.Biol., 12, 2016
|
|
8C7K
 
 | |
5D5L
 
 | |
3PH9
 
 | | Crystal structure of the human anterior gradient protein 3 | | 分子名称: | Anterior gradient protein 3 homolog | | 著者 | Nguyen, V.D, Ruddock, L.W, Salin, M, Wierenga, R.K. | | 登録日 | 2010-11-03 | | 公開日 | 2011-10-19 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Crystal structure of human anterior gradient protein 3.
Acta Crystallogr.,Sect.F, 74, 2018
|
|
8C3L
 
 | |
305D
 
 | |
306D
 
 | |
304D
 
 | |
8FE5
 
 | | Structure of J-PKAc chimera complexed with Aplithianine B | | 分子名称: | 6-[(6P)-6-(1-methyl-1H-imidazol-5-yl)-2,3-dihydro-4H-1,4-thiazin-4-yl]-7,9-dihydro-8H-purin-8-one, DnaJ homolog subfamily B member 1,cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | 著者 | Du, L, Wilson, B.A.P, Li, N, Martinez Fiesco, J.A, Dalilian, M, Wang, D, Smith, E.A, Wamiru, A, Goncharova, E.I, Zhang, P, O'Keefe, B.R. | | 登録日 | 2022-12-05 | | 公開日 | 2023-10-18 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Discovery and Synthesis of a Naturally Derived Protein Kinase Inhibitor that Selectively Inhibits Distinct Classes of Serine/Threonine Kinases.
J.Nat.Prod., 86, 2023
|
|
8G87
 
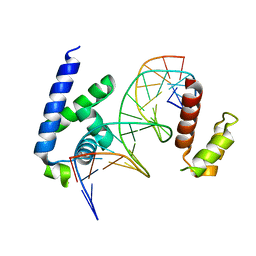 | | Human Oct4 bound to nucleosome with human nMatn1 sequence (focused refinement of Oct4 bound region) | | 分子名称: | POU domain, class 5, transcription factor 1, ... | | 著者 | Sinha, K.K, Bilokapic, S, Du, Y, Malik, D, Halic, M. | | 登録日 | 2023-02-17 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (8.1 Å) | | 主引用文献 | Histone modifications regulate pioneer transcription factor cooperativity.
Nature, 619, 2023
|
|
8G86
 
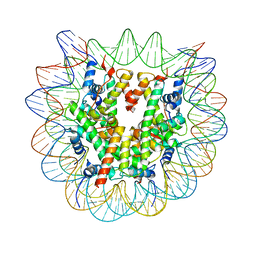 | | Human Oct4 bound to nucleosome with human nMatn1 sequence (focused refinement of nucleosome) | | 分子名称: | Histone H2A, Histone H2B, Histone H3, ... | | 著者 | Sinha, K.K, Bilokapic, S, Du, Y, Malik, D, Halic, M. | | 登録日 | 2023-02-17 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.3 Å) | | 主引用文献 | Histone modifications regulate pioneer transcription factor cooperativity.
Nature, 619, 2023
|
|
8G88
 
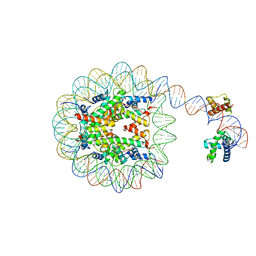 | | Human Oct4 bound to nucleosome with human nMatn1 sequence | | 分子名称: | Histone H2A, Histone H2B, Histone H3, ... | | 著者 | Sinha, K.K, Bilokapic, S, Du, Y, Malik, D, Halic, M. | | 登録日 | 2023-02-17 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.3 Å) | | 主引用文献 | Histone modifications regulate pioneer transcription factor cooperativity.
Nature, 619, 2023
|
|
8G8B
 
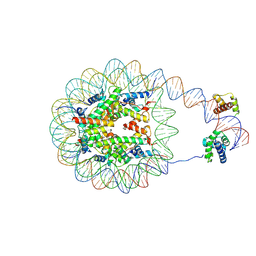 | | Nucleosome with human nMatn1 sequence in complex with Human Oct4 | | 分子名称: | Histone H2A, Histone H2B, Histone H3, ... | | 著者 | Sinha, K.K, Bilokapic, S, Du, Y, Malik, D, Halic, M. | | 登録日 | 2023-02-17 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Histone modifications regulate pioneer transcription factor cooperativity.
Nature, 619, 2023
|
|
8G8E
 
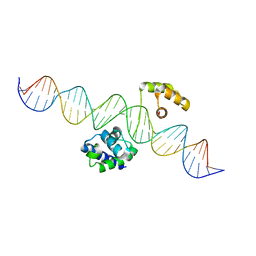 | | Human Oct4 bound to nucleosome with human LIN28B sequence | | 分子名称: | LIN28B DNA (32-MER), POU domain, class 5, ... | | 著者 | Sinha, K.K, Bilokapic, S, Du, Y, Malik, D, Halic, M. | | 登録日 | 2023-02-17 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Histone modifications regulate pioneer transcription factor cooperativity.
Nature, 619, 2023
|
|
8G8G
 
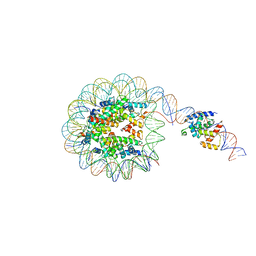 | | Interaction of H3 tail in LIN28B nucleosome with Oct4 | | 分子名称: | Histone H2A, Histone H2B, Histone H3, ... | | 著者 | Sinha, K.K, Bilokapic, S, Du, Y, Malik, D, Halic, M. | | 登録日 | 2023-02-17 | | 公開日 | 2023-03-22 | | 最終更新日 | 2023-12-13 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Histone modifications regulate pioneer transcription factor cooperativity.
Nature, 619, 2023
|
|
8BT1
 
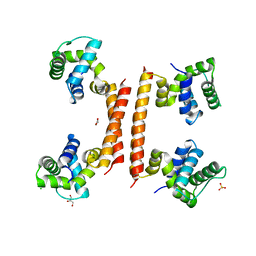 | | YdaT transcription regulator (CII functional analog) | | 分子名称: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | 著者 | Prolic-Kalinsek, M, Loris, R. | | 登録日 | 2022-11-27 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.39788437 Å) | | 主引用文献 | Structural basis of DNA binding by YdaT, a functional equivalent of the CII repressor in the cryptic prophage CP-933P from Escherichia coli O157:H7.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8CO2
 
 | |
