3B3V
 
 | | Crystal structure of the S228A mutant of the aminopeptidase from Vibrio proteolyticus | | 分子名称: | Bacterial leucyl aminopeptidase, SODIUM ION, THIOCYANATE ION, ... | | 著者 | Ataie, N.J, Hoang, Q.Q, Zahniser, M.P.D, Milne, A, Petsko, G.A, Ringe, D. | | 登録日 | 2007-10-22 | | 公開日 | 2007-11-27 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.22 Å) | | 主引用文献 | Zinc coordination geometry and ligand binding affinity: the structural and kinetic analysis of the second-shell serine 228 residue and the methionine 180 residue of the aminopeptidase from Vibrio proteolyticus.
Biochemistry, 47, 2008
|
|
3AAT
 
 | |
3B35
 
 | | Crystal structure of the M180A mutant of the aminopeptidase from Vibrio proteolyticus | | 分子名称: | Bacterial leucyl aminopeptidase, SODIUM ION, THIOCYANATE ION, ... | | 著者 | Ataie, N.J, Hoang, Q.Q, Petsko, G.A, Ringe, D. | | 登録日 | 2007-10-19 | | 公開日 | 2007-11-27 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Zinc coordination geometry and ligand binding affinity: the structural and kinetic analysis of the second-shell serine 228 residue and the methionine 180 residue of the aminopeptidase from Vibrio proteolyticus.
Biochemistry, 47, 2008
|
|
3B3T
 
 | | Crystal structure of the D118N mutant of the aminopeptidase from Vibrio proteolyticus | | 分子名称: | Bacterial leucyl aminopeptidase, ISOLEUCINE, SODIUM ION, ... | | 著者 | Ataie, N.J, Hoang, Q.Q, Zahniser, M.P.D, Milne, A, Petsko, G.A, Ringe, D. | | 登録日 | 2007-10-22 | | 公開日 | 2007-11-27 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.17 Å) | | 主引用文献 | Zinc coordination geometry and ligand binding affinity: the structural and kinetic analysis of the second-shell serine 228 residue and the methionine 180 residue of the aminopeptidase from Vibrio proteolyticus.
Biochemistry, 47, 2008
|
|
3B3C
 
 | | Crystal structure of the M180A mutant of the aminopeptidase from Vibrio proteolyticus in complex with leucine phosphonic acid | | 分子名称: | Bacterial leucyl aminopeptidase, LEUCINE PHOSPHONIC ACID, POTASSIUM ION, ... | | 著者 | Ataie, N.J, Hoang, Q.Q, Petsko, G.A, Ringe, D. | | 登録日 | 2007-10-19 | | 公開日 | 2007-11-27 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Zinc coordination geometry and ligand binding affinity: the structural and kinetic analysis of the second-shell serine 228 residue and the methionine 180 residue of the aminopeptidase from Vibrio proteolyticus.
Biochemistry, 47, 2008
|
|
3B3S
 
 | | Crystal structure of the M180A mutant of the aminopeptidase from Vibrio proteolyticus in complex with leucine | | 分子名称: | Bacterial leucyl aminopeptidase, LEUCINE, SODIUM ION, ... | | 著者 | Ataie, N.J, Hoang, Q.Q, Petsko, G.A, Ringe, D. | | 登録日 | 2007-10-22 | | 公開日 | 2007-11-27 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.18 Å) | | 主引用文献 | Zinc coordination geometry and ligand binding affinity: the structural and kinetic analysis of the second-shell serine 228 residue and the methionine 180 residue of the aminopeptidase from Vibrio proteolyticus.
Biochemistry, 47, 2008
|
|
3BBG
 
 | |
3B3W
 
 | | Crystal structure of the S228A mutant of the aminopeptidase from Vibrio proteolyticus in complex with leucine | | 分子名称: | Bacterial leucyl aminopeptidase, LEUCINE, SODIUM ION, ... | | 著者 | Ataie, N.J, Hoang, Q.Q, Zahniser, M.P.D, Milne, A, Petsko, G.A, Ringe, D. | | 登録日 | 2007-10-22 | | 公開日 | 2007-11-27 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Zinc coordination geometry and ligand binding affinity: the structural and kinetic analysis of the second-shell serine 228 residue and the methionine 180 residue of the aminopeptidase from Vibrio proteolyticus.
Biochemistry, 47, 2008
|
|
3B7I
 
 | | Crystal structure of the S228A mutant of the aminopeptidase from Vibrio proteolyticus in complex with leucine phosphonic acid | | 分子名称: | Bacterial leucyl aminopeptidase, LEUCINE, LEUCINE PHOSPHONIC ACID, ... | | 著者 | Ataie, N.J, Hoang, Q.Q, Zahniser, M.P.D, Milne, A, Petsko, G.A, Ringe, D. | | 登録日 | 2007-10-30 | | 公開日 | 2007-11-27 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Zinc coordination geometry and ligand binding affinity: the structural and kinetic analysis of the second-shell serine 228 residue and the methionine 180 residue of the aminopeptidase from Vibrio proteolyticus.
Biochemistry, 47, 2008
|
|
1MDR
 
 | | THE ROLE OF LYSINE 166 IN THE MECHANISM OF MANDELATE RACEMASE FROM PSEUDOMONAS PUTIDA: MECHANISTIC AND CRYSTALLOGRAPHIC EVIDENCE FOR STEREOSPECIFIC ALKYLATION BY (R)-ALPHA-PHENYLGLYCIDATE | | 分子名称: | ATROLACTIC ACID (2-PHENYL-LACTIC ACID), MAGNESIUM ION, MANDELATE RACEMASE | | 著者 | Landro, J.A, Gerlt, J.A, Kozarich, J.W, Koo, C.W, Shah, V.J, Kenyon, G.L, Neidhart, D.J, Fujita, S, Petsko, G.A. | | 登録日 | 1993-11-19 | | 公開日 | 1994-08-31 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The role of lysine 166 in the mechanism of mandelate racemase from Pseudomonas putida: mechanistic and crystallographic evidence for stereospecific alkylation by (R)-alpha-phenylglycidate.
Biochemistry, 33, 1994
|
|
5T1J
 
 | | Crystal Structure of the Tbox DNA binding domain of the transcription factor T-bet | | 分子名称: | DNA, T-box transcription factor TBX21 | | 著者 | Liu, C.F, Brandt, G.S, Hoang, Q, Hwang, E.S, Naumova, N, Lazarevic, V, Dekker, J, Glimcher, L.H, Ringe, D, Petsko, G.A. | | 登録日 | 2016-08-19 | | 公開日 | 2016-10-26 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.947 Å) | | 主引用文献 | Crystal structure of the DNA binding domain of the transcription factor T-bet suggests simultaneous recognition of distant genome sites.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5T4J
 
 | | PLP and GABA Trigger GabR-Mediated Transcription Regulation in Bacillus subsidies via External Aldimine Formation | | 分子名称: | GAMMA-AMINO-BUTANOIC ACID, HTH-type transcriptional regulatory protein GabR, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Wu, R, Sanishvili, R, Belitsky, B.R, Juncosa, J.I, Le, H.V, Lehrer, H.J.S, Farley, M, Silverman, R.B, Petsko, G.A, Ringe, D, Liu, D. | | 登録日 | 2016-08-29 | | 公開日 | 2017-03-29 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.231 Å) | | 主引用文献 | PLP and GABA trigger GabR-mediated transcription regulation in Bacillus subtilis via external aldimine formation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5UCD
 
 | | Benzaldehyde Dehydrogenase, a Class 3 Aldehyde Dehydrogenase, with bound NADP+ and Benzoate Adduct | | 分子名称: | NAD(P)-dependent benzaldehyde dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Zahniser, M.P.D, Prasad, S, Kneen, M.M, Kreinbring, C.A, Petsko, G.A, Ringe, D, McLeish, M.J. | | 登録日 | 2016-12-22 | | 公開日 | 2017-04-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Structure and mechanism of benzaldehyde dehydrogenase from Pseudomonas putida ATCC 12633, a member of the Class 3 aldehyde dehydrogenase superfamily.
Protein Eng. Des. Sel., 30, 2017
|
|
5UGQ
 
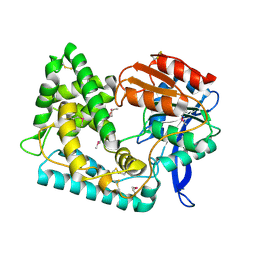 | | Crystal Structure of Hip1 (Rv2224c) | | 分子名称: | Carboxylesterase A | | 著者 | Naffin-Olivos, J.L, Daab, A, White, A, Goldfarb, N, Milne, A.C, Liu, D, Dunn, B.M, Rengarajan, J, Petsko, G.A, Ringe, D. | | 登録日 | 2017-01-09 | | 公開日 | 2017-04-12 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.609 Å) | | 主引用文献 | Structure Determination of Mycobacterium tuberculosis Serine Protease Hip1 (Rv2224c).
Biochemistry, 56, 2017
|
|
5UOH
 
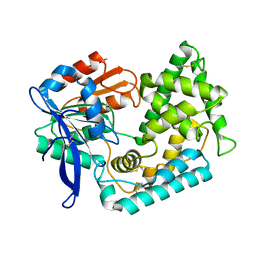 | | Crystal Structure of Hip1 (Rv2224c) T466A mutant | | 分子名称: | Carboxylesterase A | | 著者 | Naffin-Olivos, J.L, Daab, A, White, A, Goldfarb, N, Milne, A.C, Liu, D, Baikovitz, J, Dunn, B.M, Rengarajan, J, Petsko, G.A, Ringe, D. | | 登録日 | 2017-01-31 | | 公開日 | 2017-04-12 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.609 Å) | | 主引用文献 | Structure Determination of Mycobacterium tuberculosis Serine Protease Hip1 (Rv2224c).
Biochemistry, 56, 2017
|
|
5T4L
 
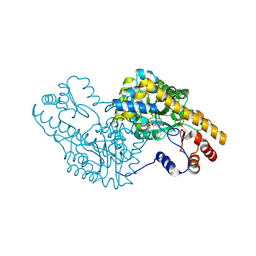 | | PLP and GABA Trigger GabR-Mediated Transcription Regulation in Bacillus subsidies via External Aldimine Formation | | 分子名称: | (4R)-4-amino-6-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}hexanoic acid, Aspartate aminotransferase | | 著者 | Wu, R, Sanishvili, R, Belitsky, B.R, Juncosa, J.I, Le, H.V, Lehrer, H.J.S, Farley, M, Silverman, R.B, Petsko, G.A, Ringe, D, Liu, D. | | 登録日 | 2016-08-29 | | 公開日 | 2017-03-29 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | PLP and GABA trigger GabR-mediated transcription regulation in Bacillus subtilis via external aldimine formation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5T4K
 
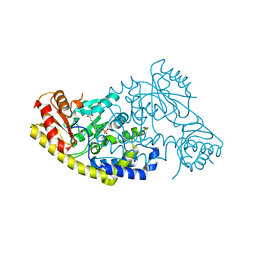 | | PLP and GABA Trigger GabR-Mediated Transcription Regulation in Bacillus subsidies via External Aldimine Formation | | 分子名称: | (4S)-5-fluoro-4-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]pentanoic acid, HTH-type transcriptional regulatory protein GabR | | 著者 | Wu, R, Sanishvili, R, Belitsky, B.R, Juncosa, J.I, Le, H.V, Lehrer, H.J.S, Farley, M, Silverman, R.B, Petsko, G.A, Ringe, D, Liu, D. | | 登録日 | 2016-08-29 | | 公開日 | 2017-03-29 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.245 Å) | | 主引用文献 | PLP and GABA trigger GabR-mediated transcription regulation in Bacillus subtilis via external aldimine formation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5UNO
 
 | | Crystal Structure of Hip1 (Rv2224c) | | 分子名称: | Carboxylesterase A | | 著者 | Naffin-Olivos, J.L, Daab, A, White, A, Goldfarb, N, Milne, A.C, Liu, D, Dunn, B.M, Rengarajan, J, Petsko, G.A, Ringe, D. | | 登録日 | 2017-01-31 | | 公開日 | 2017-04-12 | | 最終更新日 | 2019-12-11 | | 実験手法 | X-RAY DIFFRACTION (2.603 Å) | | 主引用文献 | Structure Determination of Mycobacterium tuberculosis Serine Protease Hip1 (Rv2224c).
Biochemistry, 56, 2017
|
|
4PTH
 
 | | Ensemble model for Escherichia coli dihydrofolate reductase at 100K | | 分子名称: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | 著者 | Keedy, D.A, van den Bedem, H, Sivak, D.A, Petsko, G.A, Ringe, D, Wilson, M.A, Fraser, J.S. | | 登録日 | 2014-03-10 | | 公開日 | 2014-05-14 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (0.85 Å) | | 主引用文献 | Crystal Cryocooling Distorts Conformational Heterogeneity in a Model Michaelis Complex of DHFR.
Structure, 22, 2014
|
|
4QYX
 
 | | Crystal structure of YDR533Cp | | 分子名称: | Probable chaperone protein HSP31 | | 著者 | Wilson, M.A, Amour, S.T, Collins, J.L, Ringe, D, Petsko, G.A. | | 登録日 | 2014-07-26 | | 公開日 | 2014-08-06 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | The 1.8-A resolution crystal structure of YDR533Cp from Saccharomyces cerevisiae: A member of the DJ-1/ThiJ/PfpI superfamily.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
4PST
 
 | | Multiconformer model for Escherichia coli dihydrofolate reductase at 277 K | | 分子名称: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | 著者 | Keedy, D.A, van den Bedem, H, Sivak, D.A, Petsko, G.A, Ringe, D, Wilson, M.A, Fraser, J.S. | | 登録日 | 2014-03-07 | | 公開日 | 2014-06-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Crystal Cryocooling Distorts Conformational Heterogeneity in a Model Michaelis Complex of DHFR.
Structure, 22, 2014
|
|
4PTJ
 
 | | Ensemble model for Escherichia coli dihydrofolate reductase at 277K | | 分子名称: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | 著者 | Keedy, D.A, van den Bedem, H, Sivak, D.A, Petsko, G.A, Ringe, D, Wilson, M.A, Fraser, J.S. | | 登録日 | 2014-03-10 | | 公開日 | 2014-05-14 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Crystal Cryocooling Distorts Conformational Heterogeneity in a Model Michaelis Complex of DHFR.
Structure, 22, 2014
|
|
4PSS
 
 | | Multiconformer model for Escherichia coli dihydrofolate reductase at 100K | | 分子名称: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | 著者 | Keedy, D.A, van den Bedem, H, Sivak, D.A, Petsko, G.A, Ringe, D, Wilson, M.A, Fraser, J.S. | | 登録日 | 2014-03-07 | | 公開日 | 2014-06-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (0.849 Å) | | 主引用文献 | Crystal Cryocooling Distorts Conformational Heterogeneity in a Model Michaelis Complex of DHFR.
Structure, 22, 2014
|
|
4RHN
 
 | | HISTIDINE TRIAD NUCLEOTIDE-BINDING PROTEIN (HINT) FROM RABBIT COMPLEXED WITH ADENOSINE | | 分子名称: | HISTIDINE TRIAD NUCLEOTIDE-BINDING PROTEIN, alpha-D-ribofuranose | | 著者 | Brenner, C, Garrison, P, Gilmour, J, Peisach, D, Ringe, D, Petsko, G.A, Lowenstein, J.M. | | 登録日 | 1997-02-26 | | 公開日 | 1997-06-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structures of HINT demonstrate that histidine triad proteins are GalT-related nucleotide-binding proteins.
Nat.Struct.Biol., 4, 1997
|
|
4RAT
 
 | |
