5AZ7
 
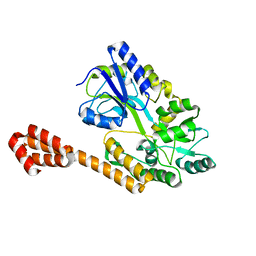 | |
5B29
 
 | | The 1.28A structure of human FABP3 F16V mutant complexed with palmitic acid at room temperature | | 分子名称: | Fatty acid-binding protein, heart, PALMITIC ACID | | 著者 | Matsuoka, D, Sugiyama, S, Kakinouchi, K, Niiyama, M, Murata, M, Matsuoka, S. | | 登録日 | 2016-01-12 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.28 Å) | | 主引用文献 | The 1.28A structure of human FABP3 F16V mutant complexed with palmitic acid at room temperature.
To Be Published
|
|
6J60
 
 | | hnRNP A1 reversible amyloid core GFGGNDNFG (residues 209-217) | | 分子名称: | 9-mer peptide (GFGGNDNFG) from Heterogeneous nuclear ribonucleoprotein A1 | | 著者 | Luo, F, Zhou, H, Gui, X, Li, D, Li, X, Liu, C. | | 登録日 | 2019-01-12 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (0.96 Å) | | 主引用文献 | Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly.
Nat Commun, 10, 2019
|
|
5AWR
 
 | |
6YKZ
 
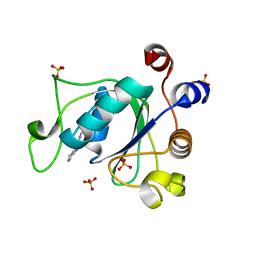 | | Crystal structure of YTHDC1 with compound DHU_DC1_234 | | 分子名称: | SULFATE ION, YTHDC1, ~{N}-methyl-1,4,5,6-tetrahydrocyclopenta[c]pyrazole-3-carboxamide | | 著者 | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | 登録日 | 2020-04-06 | | 公開日 | 2020-07-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
5B27
 
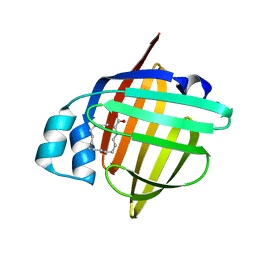 | | The 1.02A structure of human FABP3 M20S mutant complexed with palmitic acid | | 分子名称: | Fatty acid-binding protein, heart, PALMITIC ACID, ... | | 著者 | Matsuoka, D, Sugiyama, S, Kakinouchi, K, Niiyama, M, Murata, M, Matsuoka, S. | | 登録日 | 2016-01-12 | | 公開日 | 2017-01-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.02 Å) | | 主引用文献 | The 1.02A structure of human FABP3 M20S mutant complexed with palmitic acid.
To Be Published
|
|
6JB1
 
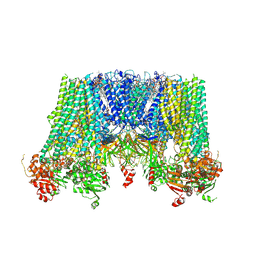 | | Structure of pancreatic ATP-sensitive potassium channel bound with repaglinide and ATPgammaS at 3.3A resolution | | 分子名称: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, ATP-binding cassette sub-family C member 8 isoform X2, ATP-sensitive inward rectifier potassium channel 11, ... | | 著者 | Chen, L, Ding, D, Wang, M, Wu, J.-X, Kang, Y. | | 登録日 | 2019-01-25 | | 公開日 | 2019-05-22 | | 最終更新日 | 2021-09-29 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | The Structural Basis for the Binding of Repaglinide to the Pancreatic KATPChannel.
Cell Rep, 27, 2019
|
|
6YKE
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_038 | | 分子名称: | (2~{R})-2-(3-fluorophenyl)-5,5-dimethyl-morpholine, SULFATE ION, YTHDC1 | | 著者 | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | 登録日 | 2020-04-06 | | 公開日 | 2020-07-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
5AQX
 
 | | Fragment-based screening of HSP70 sheds light on the functional role of ATP-binding site residues | | 分子名称: | (1R,2S,3R,5R)-3-((5-(benzyloxy)quinazolin-4-yl)amino)-5-(hydroxymethyl)cyclopentane-1,2-diol, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Jones, A.M, Westwood, I.M, Osborne, J.D, Matthews, T.P, Cheeseman, M.D, Rowlands, M.G, Jeganathan, F, Burke, R, Lee, D, Kadi, N, Liu, M, Richards, M, McAndrew, C, Yahya, N, Dobson, S.E, Jones, K, Workman, P, Collins, I, van Montfort, R.L.M. | | 登録日 | 2015-09-22 | | 公開日 | 2016-10-05 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | A fragment-based approach applied to a highly flexible target: Insights and challenges towards the inhibition of HSP70 isoforms.
Sci Rep, 6, 2016
|
|
6YL0
 
 | | Crystal structure of YTHDC1 with compound T_96 | | 分子名称: | DI(HYDROXYETHYL)ETHER, SULFATE ION, YTHDC1, ... | | 著者 | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | 登録日 | 2020-04-06 | | 公開日 | 2020-07-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Crystal structure of YTHDC1 with compound T_96
To Be Published
|
|
6YL9
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_085 | | 分子名称: | 3-[(2~{R},5~{S})-2-(2,5-dimethylphenyl)-5-methyl-morpholin-4-yl]propane-1-sulfonamide, SULFATE ION, YTHDC1 | | 著者 | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | 登録日 | 2020-04-06 | | 公開日 | 2020-07-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
5AQL
 
 | | Fragment-based screening of HSP70 sheds light on the functional role of ATP-binding site residues | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BAG FAMILY MOLECULAR CHAPERONE REGULATOR 1, GLYCEROL, ... | | 著者 | Jones, A.M, Westwood, I.M, Osborne, J.D, Matthews, T.P, Cheeseman, M.D, Rowlands, M.G, Jeganathan, F, Burke, R, Lee, D, Kadi, N, Liu, M, Richards, M, McAndrew, C, Yahya, N, Dobson, S.E, Jones, K, Workman, P, Collins, I, van Montfort, R.L.M. | | 登録日 | 2015-09-22 | | 公開日 | 2016-10-05 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | A fragment-based approach applied to a highly flexible target: Insights and challenges towards the inhibition of HSP70 isoforms.
Sci Rep, 6, 2016
|
|
1IMT
 
 | | MAMBA INTESTINAL TOXIN 1, NMR, 39 STRUCTURES | | 分子名称: | INTESTINAL TOXIN 1 | | 著者 | Boisbouvier, J, Albrand, J.-P, Blackledge, M, Jaquinod, M, Schweitz, H, Lazdunski, M, Marion, D. | | 登録日 | 1998-04-14 | | 公開日 | 1999-04-20 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A structural homologue of colipase in black mamba venom revealed by NMR floating disulphide bridge analysis.
J.Mol.Biol., 283, 1998
|
|
6J2Z
 
 | |
5B1Q
 
 | | Human herpesvirus 6B tegument protein U14 | | 分子名称: | GLYCEROL, U14 protein | | 著者 | Wang, B, Nishimura, M, Tang, H, Kawabata, A, Mahmoud, N.F, Khanlari, Z, Hamada, D, Tsuruta, H, Mori, Y. | | 登録日 | 2015-12-11 | | 公開日 | 2016-04-20 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal Structure of Human Herpesvirus 6B Tegument Protein U14.
Plos Pathog., 12, 2016
|
|
6Y3U
 
 | | Crystal structure of PPARgamma in complex with compound (R)-16 | | 分子名称: | (2~{R})-2-[[6-[(2,4-dichlorophenyl)sulfonylamino]-1,3-benzothiazol-2-yl]sulfanyl]octanoic acid, Peroxisome proliferator-activated receptor gamma | | 著者 | Chaikuad, A, Hanke, T, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2020-02-18 | | 公開日 | 2020-04-22 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.62 Å) | | 主引用文献 | A Selective Modulator of Peroxisome Proliferator-Activated Receptor gamma with an Unprecedented Binding Mode.
J.Med.Chem., 63, 2020
|
|
6Y4K
 
 | |
5AZ8
 
 | |
5B59
 
 | | Hen egg-white lysozyme modified with a keto-ABNO. | | 分子名称: | (2~{S})-2-azanyl-3-[(2~{R},3~{S})-2-oxidanyl-3-[[(1~{S},5~{R})-3-oxidanylidene-9-azabicyclo[3.3.1]nonan-9-yl]oxy]-1,2-dihydroindol-3-yl]propanal, Lysozyme C | | 著者 | Sasaki, D, Seki, Y, Sohma, Y, Oisaki, K, Kanai, M. | | 登録日 | 2016-04-28 | | 公開日 | 2016-09-14 | | 最終更新日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Transition Metal-Free Tryptophan-Selective Bioconjugation of Proteins
J.Am.Chem.Soc., 138, 2016
|
|
5AQ0
 
 | | The structure of the Transthyretin-like domain of the first catalytic domain of the HUMAN Carboxypeptidase D | | 分子名称: | CARBOXYPEPTIDASE D, GLYCEROL | | 著者 | Gallego, P, Garcia-Pardo, J, Lorenzo, J, Aviles, F.X, Ventura, S, Reverter, D. | | 登録日 | 2015-09-18 | | 公開日 | 2016-09-28 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (0.95 Å) | | 主引用文献 | The Structure of the Ttldomain of the Human Carboxypeptidase D
To be Published
|
|
4HU2
 
 | | Crystal structure of LdtMt2, a L,D-transpeptidase from Mycobacterium tuberculosis: domain A and B | | 分子名称: | PROBABLE CONSERVED LIPOPROTEIN LPPS, SULFATE ION | | 著者 | Both, D, Steiner, E, Lindqvist, Y, Schnell, R, Schneider, G. | | 登録日 | 2012-11-02 | | 公開日 | 2012-12-12 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Structure of LdtMt2, an L,D-transpeptidase from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6Y7M
 
 | | Crystal structure of the complex resulting from the reaction between the SARS-CoV main protease and tert-butyl (1-((S)-3-cyclohexyl-1-(((S)-4-(cyclopropylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)butan-2-yl)amino)-1-oxopropan-2-yl)-2-oxo-1,2-dihydropyridin-3-yl)carbamate | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclohexyl-1-[[(2~{S},3~{R})-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | 著者 | Zhang, L, Lin, D, Hilgenfeld, R. | | 登録日 | 2020-03-01 | | 公開日 | 2020-03-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors.
Science, 368, 2020
|
|
1J9Y
 
 | | Crystal structure of mannanase 26A from Pseudomonas cellulosa | | 分子名称: | MANNANASE A, ZINC ION | | 著者 | Hogg, D, Woo, E.-J, Bolam, D.N, McKie, V.A, Gilbert, H.J, Pickersgill, R.W. | | 登録日 | 2001-05-29 | | 公開日 | 2001-06-20 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal structure of mannanase 26A from Pseudomonas cellulosa and analysis of residues involved in substrate binding
J.Biol.Chem., 276, 2001
|
|
4HT3
 
 | | The crystal structure of Salmonella typhimurium Tryptophan Synthase at 1.30A complexed with N-(4'-TRIFLUOROMETHOXYBENZENESULFONYL)-2-AMINO-1-ETHYLPHOSPHATE (F9) inhibitor in the alpha site, internal aldimine | | 分子名称: | 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, ... | | 著者 | Hilario, E, Niks, D, Dunn, M.F, Mueller, L.J, Fan, L. | | 登録日 | 2012-10-31 | | 公開日 | 2014-01-01 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Allostery and substrate channeling in the tryptophan synthase bienzyme complex: evidence for two subunit conformations and four quaternary states.
Biochemistry, 52, 2013
|
|
6J8I
 
 | | Structure of human voltage-gated sodium channel Nav1.7 in complex with auxiliary beta subunits, ProTx-II and tetrodotoxin (Y1755 up) | | 分子名称: | (1R,5R,6R,7R,9S,11S,12S,13S,14S)-3-amino-14-(hydroxymethyl)-8,10-dioxa-2,4-diazatetracyclo[7.3.1.1~7,11~.0~1,6~]tetradec-3-ene-5,9,12,13,14-pentol (non-preferred name), 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Shen, H, Liu, D, Lei, J, Yan, N. | | 登録日 | 2019-01-19 | | 公開日 | 2019-02-27 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structures of human Nav1.7 channel in complex with auxiliary subunits and animal toxins.
Science, 363, 2019
|
|
