3MVL
 
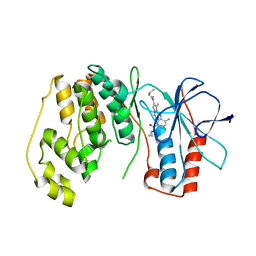 | | P38 Alpha Map Kinase complexed with pyrrolotriazine inhibitor 7K | | 分子名称: | 4-{[5-(cyclopropylcarbamoyl)-2-methylphenyl]amino}-5-methyl-N-propylpyrrolo[2,1-f][1,2,4]triazine-6-carboxamide, Mitogen-activated protein kinase 14 | | 著者 | Sack, J.S. | | 登録日 | 2010-05-04 | | 公開日 | 2010-10-13 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Discovery of 4-(5-(Cyclopropylcarbamoyl)-2-Methylphenylamino)-5-Methyl-Npropylpyrrolo[1,2-F][1,2,4] Triazine-6-Carboxamide (Bms-582949), a Clinical P38 Map Kinase Inhibitor for the Treatment of Inflammatory Diseases
J.Med.Chem., 53, 2010
|
|
3NMK
 
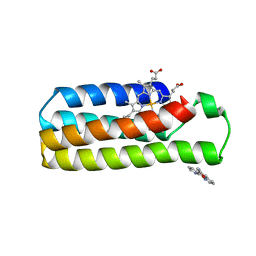 | | Crystal structure of a zinc mediated dimer for the phenanthroline-modified cytochrome cb562 variant, MBP-Phen2 | | 分子名称: | N-1,10-phenanthrolin-5-ylacetamide, PROTOPORPHYRIN IX CONTAINING FE, Soluble cytochrome b562, ... | | 著者 | Radford, R.J, Tezcan, F.A. | | 登録日 | 2010-06-22 | | 公開日 | 2010-11-24 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Porous protein frameworks with unsaturated metal centers in sterically encumbered coordination sites.
Chem.Commun.(Camb.), 47, 2011
|
|
3N7P
 
 | |
1UTR
 
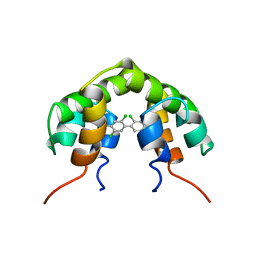 | | UTEROGLOBIN-PCB COMPLEX (REDUCED FORM) | | 分子名称: | 4,4'-BIS([H]METHYLSULFONYL)-2,2',5,5'-TETRACHLOROBIPHENYL, UTEROGLOBIN | | 著者 | Hard, T, Barnes, H.J, Larsson, C, Gustafsson, J.-A, Lund, J. | | 登録日 | 1995-09-01 | | 公開日 | 1995-12-07 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a mammalian PCB-binding protein in complex with a PCB.
Nat.Struct.Biol., 2, 1995
|
|
3Q0B
 
 | |
3Q0F
 
 | | Crystal structure of SUVH5 SRA- methylated CHH DNA complex | | 分子名称: | DNA (5'-D(*CP*TP*GP*AP*GP*GP*AP*GP*TP*AP*T)-3'), DNA (5'-D(*TP*AP*CP*TP*(5CM)P*CP*TP*CP*AP*G)-3'), Histone-lysine N-methyltransferase, ... | | 著者 | Eerappa, R, Simanshu, D.K, Patel, D.J. | | 登録日 | 2010-12-15 | | 公開日 | 2011-02-02 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo.
Genes Dev., 25, 2011
|
|
3Q32
 
 | | Structure of Janus kinase 2 with a pyrrolotriazine inhibitor | | 分子名称: | 2-(2,6-difluoro-4-methoxyphenyl)-1-(4-{4-[(3-methyl-1H-pyrazol-5-yl)amino]pyrrolo[2,1-f][1,2,4]triazin-2-yl}piperazin-1-yl)ethanone, Tyrosine-protein kinase JAK2 | | 著者 | Sack, J.S. | | 登録日 | 2010-12-21 | | 公開日 | 2011-02-16 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Pyrrolo[1,2-f]triazines as JAK2 inhibitors: Achieving potency and selectivity for JAK2 over JAK3.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3Q0C
 
 | | Crystal structure of SUVH5 SRA-fully methylated CG DNA complex in space group P6122 | | 分子名称: | DNA (5'-D(*AP*CP*TP*AP*(5CM)P*GP*TP*AP*GP*TP*T)-3'), Histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH5, ... | | 著者 | Eerappa, R, Simanshu, D.K, Patel, D.J. | | 登録日 | 2010-12-15 | | 公開日 | 2011-02-02 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.6567 Å) | | 主引用文献 | A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo.
Genes Dev., 25, 2011
|
|
1HUM
 
 | |
3PR7
 
 | |
1HR2
 
 | | CRYSTAL STRUCTURE ANALYSIS OF A MUTANT P4-P6 DOMAIN (DELC209) OF TETRAHYMENA THEMOPHILA GROUP I INTRON. | | 分子名称: | MAGNESIUM ION, P4-P6 DELC209 MUTANT RNA RIBOZYME DOMAIN | | 著者 | Juneau, K, Podell, E.R, Harrington, D.J, Cech, T.R. | | 登録日 | 2000-12-20 | | 公開日 | 2001-04-12 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structural basis of the enhanced stability of a mutant ribozyme domain and a detailed view of RNA--solvent interactions.
Structure, 9, 2001
|
|
3NTN
 
 | | Crystal Structure of UspA1 head and neck domain from Moraxella catarrhalis | | 分子名称: | CHLORIDE ION, NICKEL (II) ION, SULFATE ION, ... | | 著者 | Conners, R, Zaccai, N, Agnew, C, Burton, N, Brady, R.L. | | 登録日 | 2010-07-05 | | 公開日 | 2011-07-06 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Correlation of in situ mechanosensitive responses of the Moraxella catarrhalis adhesin UspA1 with fibronectin and receptor CEACAM1 binding.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1HSH
 
 | |
3OG7
 
 | | B-Raf Kinase V600E oncogenic mutant in complex with PLX4032 | | 分子名称: | AKAP9-BRAF fusion protein, N-(3-{[5-(4-chlorophenyl)-1H-pyrrolo[2,3-b]pyridin-3-yl]carbonyl}-2,4-difluorophenyl)propane-1-sulfonamide | | 著者 | Zhang, Y, Zhang, K.Y, Zhang, C. | | 登録日 | 2010-08-16 | | 公開日 | 2010-09-22 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma.
Nature, 467, 2010
|
|
3OHF
 
 | | Crystal structure of beta-site app-cleaving enzyme 1 (BACE-WT) complex with bms-655295 aka n~3~-((1s,2r)-1- benzyl-2-hydroxy-3-((3-methoxybenzyl)amino)propyl)-n~1~, n~1~-dibutyl-1h-indole-1,3-dicarboxamide | | 分子名称: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | 著者 | Muckelbauer, J.K. | | 登録日 | 2010-08-17 | | 公開日 | 2011-04-06 | | 最終更新日 | 2017-03-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Synthesis and SAR of indole-and 7-azaindole-1,3-dicarboxamide hydroxyethylamine inhibitors of BACE-1.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
1TAW
 
 | | BOVINE TRYPSIN COMPLEXED TO APPI | | 分子名称: | CALCIUM ION, PROTEASE INHIBITOR DOMAIN OF ALZHEIMER'S AMYLOID BETA-PROTEIN PRECURSOR, TRYPSIN | | 著者 | Hynes, T.R, Kossiakoff, A.A. | | 登録日 | 1996-12-19 | | 公開日 | 1997-06-24 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structures of bovine chymotrypsin and trypsin complexed to the inhibitor domain of Alzheimer's amyloid beta-protein precursor (APPI) and basic pancreatic trypsin inhibitor (BPTI): engineering of inhibitors with altered specificities.
Protein Sci., 6, 1997
|
|
3Q4B
 
 | | Clinically Useful Alkyl Amine Renin Inhibitors | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Renin, ... | | 著者 | Wu, Z, McKeever, B.M. | | 登録日 | 2010-12-23 | | 公開日 | 2011-11-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Discovery of VTP-27999, an Alkyl Amine Renin Inhibitor with Potential for Clinical Utility.
ACS Med Chem Lett, 2, 2011
|
|
3PWE
 
 | | Crystal structure of the E. coli beta clamp mutant R103C, I305C, C260S, C333S at 2.2A resolution | | 分子名称: | DNA polymerase III subunit beta | | 著者 | Marzahn, M.R, Robbins, A.H, McKenna, R, Bloom, L.B. | | 登録日 | 2010-12-08 | | 公開日 | 2011-10-19 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.199 Å) | | 主引用文献 | The E. coli clamp loader can actively pry open the beta-sliding clamp
J.Biol.Chem., 286, 2011
|
|
1TU2
 
 | | THE COMPLEX OF NOSTOC CYTOCHROME F AND PLASTOCYANIN DETERMIN WITH PARAMAGNETIC NMR. BASED ON THE STRUCTURES OF CYTOCHROME F AND PLASTOCYANIN, 10 STRUCTURES | | 分子名称: | Apocytochrome f, COPPER (II) ION, HEME C, ... | | 著者 | Diaz-Moreno, I, Diaz-Quintana, A, De la Rosa, M.A, Ubbink, M. | | 登録日 | 2004-06-24 | | 公開日 | 2005-03-01 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the complex between plastocyanin and cytochrome f from the cyanobacterium Nostoc sp. PCC 7119 as determined by paramagnetic NMR. The balance between electrostatic and hydrophobic interactions within the transient complex determines the relative orientation of the two proteins.
J.Biol.Chem., 280, 2005
|
|
3OPO
 
 | |
3P89
 
 | | FXR bound to a quinolinecarboxylic acid | | 分子名称: | 6-(4-{[3-(2,6-dichlorophenyl)-5-(1-methylethyl)isoxazol-4-yl]methoxy}phenyl)quinoline-2-carboxylic acid, Farnesoid X receptor, Nuclear receptor coactivator 1, ... | | 著者 | Madauss, K.P, Williams, S.P, Deaton, D.N. | | 登録日 | 2010-10-13 | | 公開日 | 2011-08-31 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Conformationally constrained farnesoid X receptor (FXR) agonists: Heteroaryl replacements of the naphthalene.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3Q79
 
 | |
1IEE
 
 | | STRUCTURE OF TETRAGONAL HEN EGG WHITE LYSOZYME AT 0.94 A FROM CRYSTALS GROWN BY THE COUNTER-DIFFUSION METHOD | | 分子名称: | CHLORIDE ION, LYSOZYME C, SODIUM ION | | 著者 | Sauter, C, Otalora, F, Gavira, J.-A, Vidal, O, Giege, R, Garcia-Ruiz, J.-M. | | 登録日 | 2001-04-09 | | 公開日 | 2001-08-08 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (0.94 Å) | | 主引用文献 | Structure of tetragonal hen egg-white lysozyme at 0.94 A from crystals grown by the counter-diffusion method.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
3QAI
 
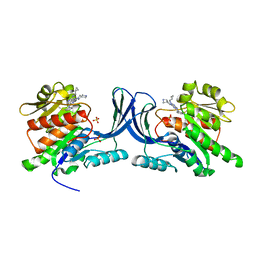 | | X-ray Structure of ketohexokinase in complex with a pyrimidopyrimidine analog 3 | | 分子名称: | Ketohexokinase, N~8~-(cyclopropylmethyl)-2-(2,6-diazaspiro[3.3]hept-2-yl)-N~4~-[2-(methylsulfanyl)phenyl]pyrimido[5,4-d]pyrimidine-4,8-diamine, SULFATE ION | | 著者 | Abad, M.C. | | 登録日 | 2011-01-11 | | 公開日 | 2012-01-18 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Inhibitors of Ketohexokinase: Discovery of Pyrimidinopyrimidines with Specific Substitution that Complements the ATP-Binding Site.
ACS Med Chem Lett, 2, 2011
|
|
3QAS
 
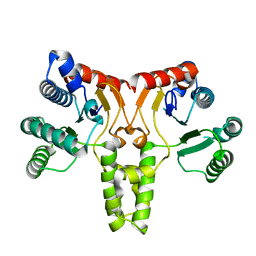 | | Structure of Undecaprenyl Diphosphate synthase | | 分子名称: | Undecaprenyl pyrophosphate synthase | | 著者 | Cao, R, Oldfield, E. | | 登録日 | 2011-01-11 | | 公開日 | 2011-03-09 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Applying Molecular Dynamics Simulations to Identify Rarely Sampled Ligand-bound Conformational States of Undecaprenyl Pyrophosphate Synthase, an Antibacterial Target.
Chem.Biol.Drug Des., 77, 2011
|
|
