8HN6
 
 | | Crystal structure of monoclonal antibody complexed with SARS-CoV-2 RBD | | 分子名称: | Heavy chain of monoclonal antibody 3G10, Light chain of monoclonal antibody 3G10, Spike protein S1 | | 著者 | Qi, J, Chen, Y. | | 登録日 | 2022-12-07 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-06-07 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Characterization of RBD-specific cross-neutralizing antibodies responses against SARS-CoV-2 variants from COVID-19 convalescents.
Front Immunol, 14, 2023
|
|
8HN7
 
 | | Crystal structure of monoclonal antibody complexed with SARS-CoV-2 RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of monoclonal antibody 3C11, Light chain of monoclonal antibody 3C11, ... | | 著者 | Qi, J, Chen, Y. | | 登録日 | 2022-12-07 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-06-07 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Characterization of RBD-specific cross-neutralizing antibodies responses against SARS-CoV-2 variants from COVID-19 convalescents.
Front Immunol, 14, 2023
|
|
6JIC
 
 | | Identification and Characterization of a carboxypeptidase inhibitor from Lycium barbarum | | 分子名称: | WCI | | 著者 | Tan, W.L, Wong, K.H, Huang, J.Y, Tay, S.V, Wang, S.J. | | 登録日 | 2019-02-20 | | 公開日 | 2020-02-26 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Identification and characterization of a wolfberry carboxypeptidase inhibitor from Lycium barbarum.
Food Chem, 351, 2021
|
|
6K13
 
 | | Crystal Structure Basis for BmLDH Complex | | 分子名称: | L-lactate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, OXAMIC ACID | | 著者 | Long, Y, Shen, Z. | | 登録日 | 2019-05-09 | | 公開日 | 2019-10-16 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Crystal structures ofBabesia microtilactate dehydrogenase BmLDH reveal a critical role for Arg99 in catalysis.
Faseb J., 33, 2019
|
|
5ID6
 
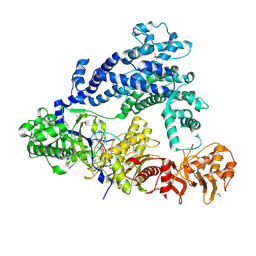 | | Structure of Cpf1/RNA Complex | | 分子名称: | Cpf1, MAGNESIUM ION, RNA (5'-R(P*AP*AP*UP*UP*UP*CP*UP*AP*CP*UP*AP*AP*GP*UP*GP*UP*AP*GP*AP*UP*C)-3') | | 著者 | Dong, D, Ren, K, Qiu, X, Wang, J, Huang, Z. | | 登録日 | 2016-02-24 | | 公開日 | 2016-04-27 | | 最終更新日 | 2016-05-11 | | 実験手法 | X-RAY DIFFRACTION (2.382 Å) | | 主引用文献 | The crystal structure of Cpf1 in complex with CRISPR RNA
Nature, 532, 2016
|
|
6DL2
 
 | | BRD4 bromodomain 1 in complex with HYB157 | | 分子名称: | 1,2-ETHANEDIOL, 3-benzyl-2,9-dimethyl-4H,6H-thieno[2,3-e][1,2,4]triazolo[3,4-c][1,4]oxazepine, Bromodomain-containing protein 4 | | 著者 | Meagher, J.L, Stuckey, J.A. | | 登録日 | 2018-05-31 | | 公開日 | 2019-04-17 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Discovery of QCA570 as an Exceptionally Potent and Efficacious Proteolysis Targeting Chimera (PROTAC) Degrader of the Bromodomain and Extra-Terminal (BET) Proteins Capable of Inducing Complete and Durable Tumor Regression.
J. Med. Chem., 61, 2018
|
|
6K12
 
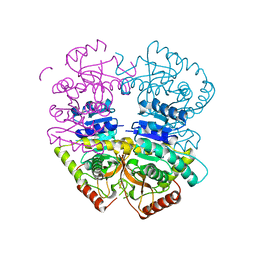 | |
8SLO
 
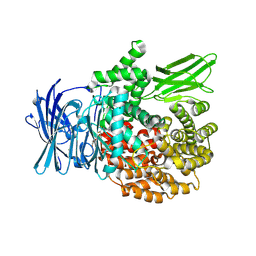 | | Plasmodium falciparum M1 aminopeptidase bound to selective inhibitor MIPS2673 | | 分子名称: | 2-hydroxy-N-[(1R)-2-(hydroxyamino)-2-oxo-1-(3',4',5'-trifluoro[1,1'-biphenyl]-4-yl)ethyl]-2-methylpropanamide, GLYCEROL, M1 family aminopeptidase, ... | | 著者 | McGowan, S.M, Drinkwater, N. | | 登録日 | 2023-04-24 | | 公開日 | 2024-04-03 | | 最終更新日 | 2024-07-24 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Chemoproteomics validates selective targeting of Plasmodium M1 alanyl aminopeptidase as an antimalarial strategy.
Elife, 13, 2024
|
|
8FP3
 
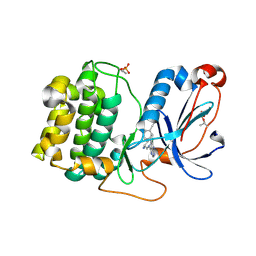 | | PKCeta kinase domain in complex with compound 11 | | 分子名称: | (3P)-3-{4-[(3R,5S)-3-amino-5-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile, Protein kinase C eta type | | 著者 | Johnson, E. | | 登録日 | 2023-01-03 | | 公開日 | 2023-04-05 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
8FH4
 
 | | Crystal structure of HPK1 kinase domain T165E,S171E phosphomimetic mutant in complex with 3-[6-chloro-4-(9-methyl-1-oxa-4,9-diazaspiro[5.5]undec-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile | | 分子名称: | (3P)-3-[6-chloro-4-(9-methyl-1-oxa-4,9-diazaspiro[5.5]undecan-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile, Mitogen-activated protein kinase kinase kinase kinase 1, SULFATE ION | | 著者 | McTigue, M, Johnson, E, Cronin, C. | | 登録日 | 2022-12-13 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.827 Å) | | 主引用文献 | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
8FKO
 
 | | Crystal structure of HPK1 kinase domain T165E,S171E phosphomimetic mutant in complex with 3-{4-[(2S,5R)-5-Amino-2-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile | | 分子名称: | (3P)-3-{4-[(2S,5R)-5-amino-2-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile, Mitogen-activated protein kinase kinase kinase kinase 1 | | 著者 | McTigue, M, Johnson, E, Cronin, C. | | 登録日 | 2022-12-21 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.104 Å) | | 主引用文献 | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
8FJZ
 
 | | Crystal structure of HPK1 kinase domain T165E,S171E phosphomimetic mutant in complex with 3-{4-[(3R,5S)-3-Amino-5-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile | | 分子名称: | (3P)-3-{4-[(3R,5S)-3-amino-5-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile, Mitogen-activated protein kinase kinase kinase kinase 1 | | 著者 | McTigue, M, Johnson, E, Cronin, C. | | 登録日 | 2022-12-20 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.897 Å) | | 主引用文献 | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
8FP1
 
 | | PKCeta kinase domain in complex with compound 2 | | 分子名称: | (3P)-3-[6-chloro-4-(9-methyl-1-oxa-4,9-diazaspiro[5.5]undecan-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile, Protein kinase C eta type | | 著者 | Johnson, E. | | 登録日 | 2023-01-03 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-04-26 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
6KMH
 
 | | The crystal structure of CASK/Mint1 complex | | 分子名称: | Amyloid-beta A4 precursor protein-binding family A member 1, CHLORIDE ION, IODIDE ION, ... | | 著者 | Li, W, Feng, W. | | 登録日 | 2019-07-31 | | 公開日 | 2020-08-05 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | CASK modulates the assembly and function of the Mint1/Munc18-1 complex to regulate insulin secretion.
Cell Discov, 6, 2020
|
|
6XM0
 
 | | Consensus structure of SARS-CoV-2 spike at pH 5.5 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhou, T, Tsybovsky, Y, Olia, A, Kwong, P.D. | | 登録日 | 2020-06-29 | | 公開日 | 2020-08-12 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
6XM5
 
 | | Structure of SARS-CoV-2 spike at pH 5.5, all RBDs down | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhou, T, Tsybovsky, Y, Olia, A, Kwong, P.D. | | 登録日 | 2020-06-29 | | 公開日 | 2020-07-29 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
6OSY
 
 | |
6XLU
 
 | | Structure of SARS-CoV-2 spike at pH 4.0 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhou, T, Tsybovsky, Y, Olia, A, Kwong, P.D. | | 登録日 | 2020-06-29 | | 公開日 | 2020-08-12 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (2.4 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
4WVD
 
 | | Identification of a novel FXR ligand that regulates metabolism | | 分子名称: | (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside, Bile acid receptor, FORMIC ACID, ... | | 著者 | Wang, R, Li, Y. | | 登録日 | 2014-11-05 | | 公開日 | 2015-02-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | The antiparasitic drug ivermectin is a novel FXR ligand that regulates metabolism.
Nat Commun, 4, 2013
|
|
6OT1
 
 | |
6XM4
 
 | | Structure of SARS-CoV-2 spike at pH 5.5, single RBD up, conformation 2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhou, T, Tsybovsky, Y, Olia, A, Kwong, P.D. | | 登録日 | 2020-06-29 | | 公開日 | 2020-08-12 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7M4T
 
 | | Menin bound to M-1121 | | 分子名称: | Menin, methyl {(1S,2R)-2-[(1S)-2-(azetidin-1-yl)-1-(3-fluorophenyl)-1-{1-[(3-methoxy-1-{4-[(1S,4S)-5-propanoyl-2,5-diazabicyclo[2.2.1]heptane-2-sulfonyl]phenyl}azetidin-3-yl)methyl]piperidin-4-yl}ethyl]cyclopentyl}carbamate, praseodymium triacetate | | 著者 | Stuckey, J. | | 登録日 | 2021-03-22 | | 公開日 | 2021-08-11 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.74 Å) | | 主引用文献 | Discovery of M-1121 as an Orally Active Covalent Inhibitor of Menin-MLL Interaction Capable of Achieving Complete and Long-Lasting Tumor Regression.
J.Med.Chem., 64, 2021
|
|
6XM3
 
 | | Structure of SARS-CoV-2 spike at pH 5.5, single RBD up, conformation 1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhou, T, Tsybovsky, Y, Olia, A, Kwong, P.D. | | 登録日 | 2020-06-29 | | 公開日 | 2020-08-12 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
5I29
 
 | | TAF1(2) bound to a pyrrolopyridone compound | | 分子名称: | CALCIUM ION, N,N-dimethyl-3-(6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl)benzamide, Transcription initiation factor TFIID subunit 1 | | 著者 | Tang, Y, Poy, F, Bellon, S.F. | | 登録日 | 2016-02-08 | | 公開日 | 2016-06-08 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.21 Å) | | 主引用文献 | Diving into the Water: Inducible Binding Conformations for BRD4, TAF1(2), BRD9, and CECR2 Bromodomains.
J.Med.Chem., 59, 2016
|
|
6W7G
 
 | | Structure of EED bound to inhibitor 1056 | | 分子名称: | 8-(2,6-dimethylpyridin-3-yl)-N-[(5-fluoro-2,3-dihydro-1-benzofuran-4-yl)methyl]-1-(methylsulfonyl)imidazo[1,5-c]pyrimidin-5-amine, FORMIC ACID, Polycomb protein EED, ... | | 著者 | Petrunak, E.M, Stuckey, J.A. | | 登録日 | 2020-03-19 | | 公開日 | 2020-07-08 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | EEDi-5285: An Exceptionally Potent, Efficacious, and Orally Active Small-Molecule Inhibitor of Embryonic Ectoderm Development.
J.Med.Chem., 63, 2020
|
|
