2K2O
 
 | |
1BOO
 
 | | PVUII DNA METHYLTRANSFERASE (CYTOSINE-N4-SPECIFIC) | | 分子名称: | PROTEIN (N-4 CYTOSINE-SPECIFIC METHYLTRANSFERASE PVU II), S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Gong, W, O'Gara, M, Blumenthal, R.M, Cheng, X. | | 登録日 | 1998-07-31 | | 公開日 | 1998-08-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of pvu II DNA-(cytosine N4) methyltransferase, an example of domain permutation and protein fold assignment.
Nucleic Acids Res., 25, 1997
|
|
3WDZ
 
 | | Crystal Structure of Keap1 in Complex with phosphorylated p62 | | 分子名称: | Kelch-like ECH-associated protein 1, Peptide from Sequestosome-1 | | 著者 | Fukutomi, T, Takagi, K, Mizushima, T, Tanaka, K, Komatsu, M, Yamamoto, M. | | 登録日 | 2013-06-26 | | 公開日 | 2013-09-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy.
Mol.Cell, 51, 2013
|
|
2DYJ
 
 | | Crystal structure of ribosome-binding factor A from Thermus thermophilus HB8 | | 分子名称: | Ribosome-binding factor A | | 著者 | Kawazoe, M, Takemoto, C, Nakayama-Ushikoshi, R, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2006-09-14 | | 公開日 | 2007-03-14 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Structural aspects of RbfA action during small ribosomal subunit assembly.
Mol.Cell, 28, 2007
|
|
5Y33
 
 | |
3VLF
 
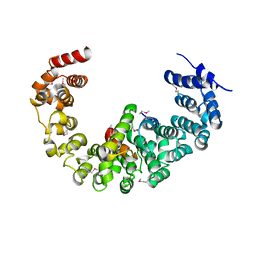 | | Crystal structure of yeast proteasome interacting protein | | 分子名称: | 26S protease regulatory subunit 7 homolog, DNA mismatch repair protein HSM3 | | 著者 | Takagi, K, Kim, S, Kato, K, Tanaka, K, Saeki, Y, Mizushima, T. | | 登録日 | 2011-12-01 | | 公開日 | 2012-02-22 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Structural basis for specific recognition of Rpt1, an ATPase subunit of the 26S proteasome, by a proteasome-dedicated chaperone Hsm3
J.Biol.Chem., 287, 2012
|
|
3VOC
 
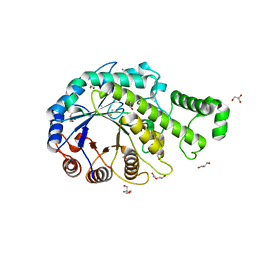 | | Crystal structure of the catalytic domain of beta-amylase from paenibacillus polymyxa | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta/alpha-amylase, ... | | 著者 | Nishimura, S, Fujioka, T, Nakaniwa, T, Tada, T. | | 登録日 | 2012-01-21 | | 公開日 | 2013-02-20 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural analysis by X-ray crystallography and small-angle scattering of the multi-domain beta-amylase from Paenibacillus polymyxa
To be Published
|
|
3VLE
 
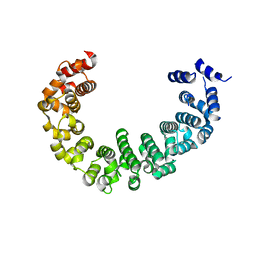 | | Crystal structure of yeast proteasome interacting protein | | 分子名称: | DNA mismatch repair protein HSM3 | | 著者 | Takagi, K, Kim, S, Kato, K, Tanaka, K, Saeki, Y, Mizushima, T. | | 登録日 | 2011-12-01 | | 公開日 | 2012-02-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Structural basis for specific recognition of Rpt1, an ATPase subunit of the 26S proteasome, by a proteasome-dedicated chaperone Hsm3
J.Biol.Chem., 287, 2012
|
|
3VLD
 
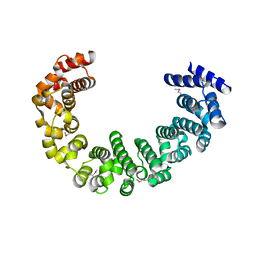 | | Crystal structure of yeast proteasome interacting protein | | 分子名称: | DNA mismatch repair protein HSM3 | | 著者 | Takagi, K, Kim, S, Kato, K, Tanaka, K, Saeki, Y, Mizushima, T. | | 登録日 | 2011-12-01 | | 公開日 | 2012-02-22 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structural basis for specific recognition of Rpt1, an ATPase subunit of the 26S proteasome, by a proteasome-dedicated chaperone Hsm3
J.Biol.Chem., 287, 2012
|
|
2L4J
 
 | | Yap ww2 | | 分子名称: | Yes-associated protein 2 (YAP2) | | 著者 | Webb, C, Upadhyay, A, Furutani-Seiki, M, Bagby, S. | | 登録日 | 2010-10-07 | | 公開日 | 2010-11-03 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Features and Ligand Binding Properties of Tandem WW Domains from YAP and TAZ, Nuclear Effectors of the Hippo Pathway.
Biochemistry, 50, 2011
|
|
2MWO
 
 | |
2K8F
 
 | | Structural Basis for the Regulation of p53 Function by p300 | | 分子名称: | Cellular tumor antigen p53, Histone acetyltransferase p300 | | 著者 | Bai, Y, Feng, H, Jenkins, L.M, Durell, S.R, Wiodawer, A, Appella, E. | | 登録日 | 2008-09-08 | | 公開日 | 2009-03-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Basis for p300 Taz2-p53 TAD1 Binding and Modulation by Phosphorylation.
Structure, 17, 2009
|
|
2ZE4
 
 | | Crystal structure of phospholipase D from streptomyces antibioticus | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Phospholipase D | | 著者 | Suzuki, A, Kakuno, K, Saito, R, Iwasaki, Y, Yamane, T, Yamane, T. | | 登録日 | 2007-12-05 | | 公開日 | 2007-12-25 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of phospholipase D from streptomyces antibioticus
To be Published
|
|
2MWP
 
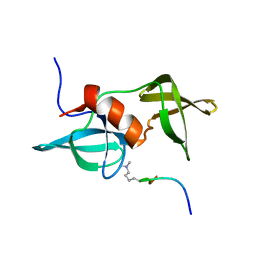 | |
2ZTN
 
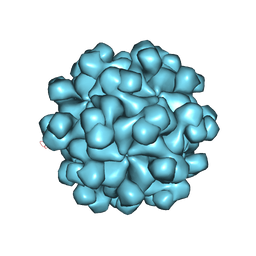 | | Hepatitis E virus ORF2 (Genotype 3) | | 分子名称: | Capsid protein | | 著者 | Yamashita, T, Unno, H, Mori, Y, Li, T.C, Takeda, N, Matsuura, Y. | | 登録日 | 2008-10-08 | | 公開日 | 2009-08-25 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.56 Å) | | 主引用文献 | Biological and immunological characteristics of hepatitis E virus-like particles based on the crystal structure
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2W1B
 
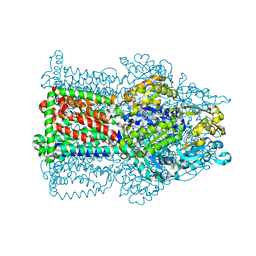 | | The structure of the efflux pump AcrB in complex with bile acid | | 分子名称: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, ACRIFLAVIN RESISTANCE PROTEIN B | | 著者 | Drew, D, Klepsch, M.M, Newstead, S, Flaig, R, De Gier, J.W, Iwata, S, Beis, K. | | 登録日 | 2008-10-17 | | 公開日 | 2008-12-09 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (3.85 Å) | | 主引用文献 | The Structure of the Efflux Pump Acrb in Complex with Bile Acid.
Mol.Membr.Biol., 25, 2008
|
|
2ZFA
 
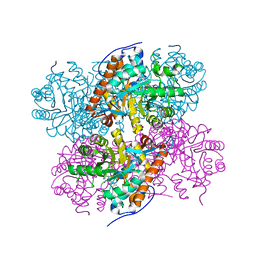 | | Structure of Lactate Oxidase at pH4.5 from AEROCOCCUS VIRIDANS | | 分子名称: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, Lactate oxidase | | 著者 | Furuichi, M, Balasundaresan, D, Suzuki, N, Yoshida, Y, Minagawa, H, Kaneko, H, Waga, I, Kumar, P.K.R, Mizuno, H. | | 登録日 | 2007-12-26 | | 公開日 | 2008-04-22 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | X-ray structures of Aerococcus viridans lactate oxidase and its complex with D-lactate at pH 4.5 show an alpha-hydroxyacid oxidation mechanism
J.Mol.Biol., 378, 2008
|
|
5T8Q
 
 | |
5T8F
 
 | | p110delta/p85alpha with taselisib (GDC-0032) | | 分子名称: | 2-methyl-2-(4-{2-[3-methyl-1-(propan-2-yl)-1H-1,2,4-triazol-5-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}-1H-pyrazol-1-yl)propanamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | 著者 | Moertl, M, Steinbacher, S, Eigenbrot, C. | | 登録日 | 2016-09-07 | | 公開日 | 2017-01-11 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.91 Å) | | 主引用文献 | Structure-Based Design of Tricyclic NF-kappa B Inducing Kinase (NIK) Inhibitors That Have High Selectivity over Phosphoinositide-3-kinase (PI3K).
J. Med. Chem., 60, 2017
|
|
5T8O
 
 | | Crystal structure of murine NF-kappaB inducing kinase (NIK) bound to Imidazobenzoxepin Compound 3 | | 分子名称: | 10-(3-methyl-3-oxidanyl-but-1-ynyl)-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepine-2-carboxamide, Mitogen-activated protein kinase kinase kinase 14, SULFATE ION | | 著者 | Smith, M.A, McEwan, P, Hymowitz, S.G. | | 登録日 | 2016-09-08 | | 公開日 | 2017-01-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Structure-Based Design of Tricyclic NF-kappa B Inducing Kinase (NIK) Inhibitors That Have High Selectivity over Phosphoinositide-3-kinase (PI3K).
J. Med. Chem., 60, 2017
|
|
5T8P
 
 | | Crystal structure of murine NF-kappaB inducing kinase (NIK) bound to benzoxepin compound 2 | | 分子名称: | 6,7-dihydrothieno[4,5]oxepino[1,2-~{c}]pyridine-2-carboxamide, Mitogen-activated protein kinase kinase kinase 14, SULFATE ION | | 著者 | Smith, M.A, McEwan, P.A. | | 登録日 | 2016-09-08 | | 公開日 | 2017-01-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Structure-Based Design of Tricyclic NF-kappa B Inducing Kinase (NIK) Inhibitors That Have High Selectivity over Phosphoinositide-3-kinase (PI3K).
J. Med. Chem., 60, 2017
|
|
2CNP
 
 | |
2ZE9
 
 | | Crystal structure of H168A mutant of phospholipase D from Streptomyces antibioticus, as a complex with phosphatidylcholine | | 分子名称: | (2R)-3-(phosphonooxy)propane-1,2-diyl diheptanoate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Phospholipase D | | 著者 | Suzuki, A, Toda, H, Iwasaki, Y, Yamane, T, Yamane, T. | | 登録日 | 2007-12-06 | | 公開日 | 2007-12-25 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of phospholipase D from streptomyces antibioticus
To be Published
|
|
3VZQ
 
 | |
3VZP
 
 | | Crystal structure of PhaB from Ralstonia eutropha | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, Acetoacetyl-CoA reductase, GLYCEROL, ... | | 著者 | Ikeda, K, Tanaka, Y, Tanaka, I, Yao, M. | | 登録日 | 2012-10-15 | | 公開日 | 2013-08-28 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.792 Å) | | 主引用文献 | Directed evolution and structural analysis of NADPH-dependent Acetoacetyl Coenzyme A (Acetoacetyl-CoA) reductase from Ralstonia eutropha reveals two mutations responsible for enhanced kinetics
Appl.Environ.Microbiol., 79, 2013
|
|
