3WQ9
 
 | | Crystal structure of Hsp90-alpha N-terminal domain in complex with 2-(4-Hydroxy-cyclohexylamino)-4-[5-(4-phenyl-imidazol-1-yl)-isoquinolin-1-yl]-benzamide | | 分子名称: | 2-[(trans-4-hydroxycyclohexyl)amino]-4-[5-(4-phenyl-1H-imidazol-1-yl)isoquinolin-1-yl]benzamide, Heat shock protein HSP 90-alpha | | 著者 | Chong, K.T, Yamashita, S, Oshiumi, H, Uno, T, Kitade, M. | | 登録日 | 2014-01-23 | | 公開日 | 2015-02-25 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Evolution of highly selective Hsp90 / inhibitors by structure and thermodynamics guided design
To be Published
|
|
1RDC
 
 | |
7EQ9
 
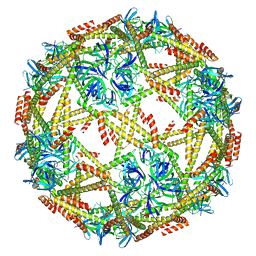 | | Cryo-EM structure of designed protein nanoparticle TIP60 (Truncated Icosahedral Protein composed of 60-mer fusion proteins) | | 分子名称: | TIP60 | | 著者 | Obata, J, Kawakami, N, Tsutsumi, A, Miyamoto, K, Kikkawa, M, Arai, R. | | 登録日 | 2021-04-30 | | 公開日 | 2021-09-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Icosahedral 60-meric porous structure of designed supramolecular protein nanoparticle TIP60.
Chem.Commun.(Camb.), 57, 2021
|
|
1RDA
 
 | |
4G78
 
 | |
6IOY
 
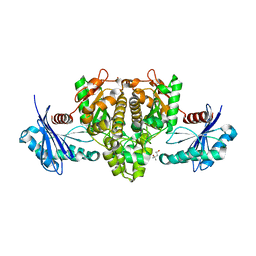 | | Crystal structure of Porphyromonas gingivalis acetate kinase | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Acetate kinase, SULFATE ION | | 著者 | Kezuka, Y, Yoshida, Y, Nonaka, T. | | 登録日 | 2018-10-31 | | 公開日 | 2019-04-24 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Characterization of the phosphotransacetylase-acetate kinase pathway for ATP production inPorphyromonas gingivalis.
J Oral Microbiol, 11, 2019
|
|
6IOW
 
 | |
5XM1
 
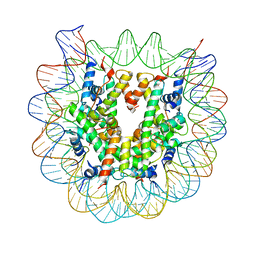 | | The mouse nucleosome structure containing H2A, H2B type3-A, H3mm7, and H4 | | 分子名称: | DNA (146-MER), Histone H2A type 1-B, Histone H2B type 3-A, ... | | 著者 | Taguchi, H, Horikoshi, N, Kurumizaka, H. | | 登録日 | 2017-05-12 | | 公開日 | 2018-03-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.45 Å) | | 主引用文献 | Histone H3.3 sub-variant H3mm7 is required for normal skeletal muscle regeneration.
Nat Commun, 9, 2018
|
|
6IOX
 
 | |
7Y3L
 
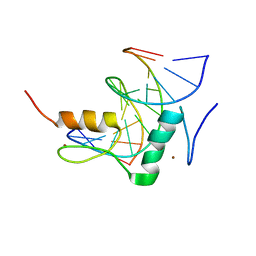 | | Structure of SALL3 ZFC4 bound with 12 bp AT-rich dsDNA | | 分子名称: | DNA (12-mer), Sal-like protein 3, ZINC ION | | 著者 | Ru, W, Xu, C. | | 登録日 | 2022-06-11 | | 公開日 | 2022-10-26 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural studies of SALL family protein zinc finger cluster domains in complex with DNA reveal preferential binding to an AATA tetranucleotide motif.
J.Biol.Chem., 298, 2022
|
|
7Y3I
 
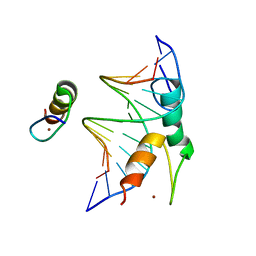 | | Structure of DNA bound SALL4 | | 分子名称: | DNA (12-mer), Sal-like protein 4, ZINC ION | | 著者 | Ru, W, Xu, C. | | 登録日 | 2022-06-10 | | 公開日 | 2022-10-26 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Structural studies of SALL family protein zinc finger cluster domains in complex with DNA reveal preferential binding to an AATA tetranucleotide motif.
J.Biol.Chem., 298, 2022
|
|
7Y3K
 
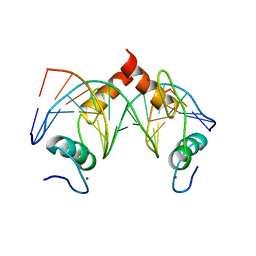 | | Structure of SALL4 ZFC4 bound with 16 bp AT-rich dsDNA | | 分子名称: | DNA (16-mer), Sal-like protein 4, ZINC ION | | 著者 | Ru, W, Xu, C. | | 登録日 | 2022-06-11 | | 公開日 | 2022-10-26 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.501 Å) | | 主引用文献 | Structural studies of SALL family protein zinc finger cluster domains in complex with DNA reveal preferential binding to an AATA tetranucleotide motif.
J.Biol.Chem., 298, 2022
|
|
7Y3M
 
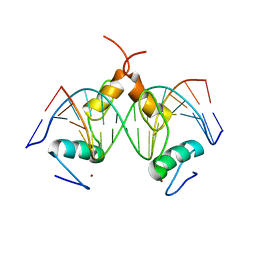 | | Structure of SALL4 ZFC1 bound with 16 bp AT-rich dsDNA | | 分子名称: | DNA (16-mer), Sal-like protein 4, ZINC ION | | 著者 | Ru, W, Xu, C. | | 登録日 | 2022-06-11 | | 公開日 | 2022-10-26 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.723 Å) | | 主引用文献 | Structural studies of SALL family protein zinc finger cluster domains in complex with DNA reveal preferential binding to an AATA tetranucleotide motif.
J.Biol.Chem., 298, 2022
|
|
5WQJ
 
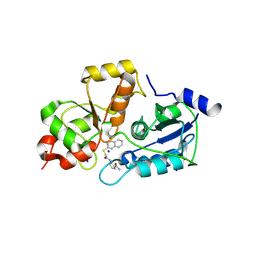 | |
5WQK
 
 | |
3GWD
 
 | |
5XM0
 
 | | The mouse nucleosome structure containing H2A, H2B type3-A, H3.3, and H4 | | 分子名称: | DNA (146-MER), Histone H2A type 1-B, Histone H2B type 3-A, ... | | 著者 | Taguchi, H, Horikoshi, N, Kurumizaka, H. | | 登録日 | 2017-05-12 | | 公開日 | 2018-03-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.874 Å) | | 主引用文献 | Histone H3.3 sub-variant H3mm7 is required for normal skeletal muscle regeneration.
Nat Commun, 9, 2018
|
|
3GWF
 
 | |
1VFH
 
 | | Crystal structure of alanine racemase from D-cycloserine producing Streptomyces lavendulae | | 分子名称: | PYRIDOXAL-5'-PHOSPHATE, alanine racemase | | 著者 | Noda, M, Matoba, Y, Kumagai, T, Sugiyama, M. | | 登録日 | 2004-04-13 | | 公開日 | 2004-09-14 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural evidence that alanine racemase from a D-cycloserine-producing microorganism exhibits resistance to its own product.
J.Biol.Chem., 279, 2004
|
|
1VFS
 
 | | Crystal structure of D-cycloserine-bound form of alanine racemase from D-cycloserine-producing Streptomyces lavendulae | | 分子名称: | CHLORIDE ION, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE, alanine racemase | | 著者 | Noda, M, Matoba, Y, Kumagai, T, Sugiyama, M. | | 登録日 | 2004-04-19 | | 公開日 | 2004-09-14 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural evidence that alanine racemase from a D-cycloserine-producing microorganism exhibits resistance to its own product.
J.Biol.Chem., 279, 2004
|
|
4XB2
 
 | | Hyperthermophilic archaeal homoserine dehydrogenase mutant in complex with NADPH | | 分子名称: | 319aa long hypothetical homoserine dehydrogenase, L-HOMOSERINE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Sakuraba, H, Inoue, S, Yoneda, K, Ohshima, T. | | 登録日 | 2014-12-16 | | 公開日 | 2015-07-15 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.43 Å) | | 主引用文献 | Crystal Structures of a Hyperthermophilic Archaeal Homoserine Dehydrogenase Suggest a Novel Cofactor Binding Mode for Oxidoreductases.
Sci Rep, 5, 2015
|
|
4XB1
 
 | | Hyperthermophilic archaeal homoserine dehydrogenase in complex with NADPH | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 319aa long hypothetical homoserine dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Sakuraba, H, Inoue, S, Yoneda, K, Ohshima, T. | | 登録日 | 2014-12-16 | | 公開日 | 2015-07-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal Structures of a Hyperthermophilic Archaeal Homoserine Dehydrogenase Suggest a Novel Cofactor Binding Mode for Oxidoreductases.
Sci Rep, 5, 2015
|
|
3VU8
 
 | | Metionyl-tRNA synthetase from Thermus thermophilus complexed with methionyl-adenylate analogue | | 分子名称: | Methionine--tRNA ligase, N-[METHIONYL]-N'-[ADENOSYL]-DIAMINOSULFONE, ZINC ION | | 著者 | Konno, M, Kato-Murayama, M, Toma-Fukai, S, Uchikawa, E, Nureki, O, Yokoyama, S. | | 登録日 | 2012-06-22 | | 公開日 | 2013-06-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The modeling of structures of specific conformation of homosysteine-AMP leading to thiolactone-formation on class Ia aminoacyl-tRNA synthetases
To be Published
|
|
3WPN
 
 | |
8GQ9
 
 | |
