7VYP
 
 | | The structure of GdmN complex with the natural tetrahedral intermediate, carbamoylated derivative, and AMP | | 分子名称: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, FE (III) ION, ... | | 著者 | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | 登録日 | 2021-11-15 | | 公開日 | 2022-11-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.88 Å) | | 主引用文献 | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
7VZY
 
 | | The structure of GdmN complex with AMP and 20-O-methyl-19-chloroproansamitocin | | 分子名称: | (5~{S},6~{E},8~{S},9~{S},12~{R},15~{E})-21-chloranyl-12,20-dimethoxy-6,8,16-trimethyl-5,9-bis(oxidanyl)-2-azabicyclo[16.3.1]docosa-1(21),6,15,18(22),19-pentaene-3,11-dione, 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, ... | | 著者 | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | 登録日 | 2021-11-17 | | 公開日 | 2022-11-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
7VYJ
 
 | | The structure of GdmN in complex with carbamoyl adenylate intermediate | | 分子名称: | 1,2-ETHANEDIOL, 5'-O-[(S)-(carbamoyloxy)(hydroxy)phosphoryl]adenosine, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | 登録日 | 2021-11-14 | | 公開日 | 2022-11-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
7VYO
 
 | | The structure of GdmN | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | 登録日 | 2021-11-14 | | 公開日 | 2022-11-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
7VZZ
 
 | | The structure of GdmN in complex with the natural tetrahedral intermediate, carbamoyl adenylate, and 20-O-methyl-19-chloroproansamitocin | | 分子名称: | (5~{S},6~{E},8~{S},9~{S},12~{R},15~{E})-21-chloranyl-12,20-dimethoxy-6,8,16-trimethyl-5,9-bis(oxidanyl)-2-azabicyclo[16.3.1]docosa-1(21),6,15,18(22),19-pentaene-3,11-dione, 1,2-ETHANEDIOL, 5'-O-[(S)-(carbamoyloxy)(hydroxy)phosphoryl]adenosine, ... | | 著者 | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | 登録日 | 2021-11-17 | | 公開日 | 2022-11-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
5V4X
 
 | | Human glucokinase in complex with novel pyrazole activator. | | 分子名称: | (2S)-3-cyclohexyl-2-[4-(cyclopentylsulfonyl)-2-oxopyridin-1(2H)-yl]-N-(1,3-thiazol-2-yl)propanamide, Glucokinase, IODIDE ION, ... | | 著者 | Skene, R.J, Hosfiled, D.J. | | 登録日 | 2017-03-10 | | 公開日 | 2017-05-31 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Discovery of potent and orally active 1,4-disubstituted indazoles as novel allosteric glucokinase activators.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
7KVZ
 
 | | Structure of hSTING in complex with novel carbocyclic pyrimidine CDN-2 | | 分子名称: | (2R,5R,7R,8R,10S,12aR,14R,15aS,16R)-7-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-2,10,16-trihydroxy-14-[(pyrimidin-4-yl)oxy]decahydro-2H,10H-5,8-methano-2lambda~5~,10lambda~5~-cyclopenta[l][1,3,6,9,11,2,10]pentaoxadiphosphacyclotetradecine-2,10-dione, Stimulator of interferon genes protein | | 著者 | Skene, R.J. | | 登録日 | 2020-11-29 | | 公開日 | 2022-02-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Identification of Novel Carbocyclic Pyrimidine Cyclic Dinucleotide STING Agonists for Antitumor Immunotherapy Using Systemic Intravenous Route.
J.Med.Chem., 64, 2021
|
|
1DJX
 
 | | PHOSPHOINOSITIDE-SPECIFIC PHOSPHOLIPASE C-DELTA1 FROM RAT COMPLEXED WITH INOSITOL-1,4,5-TRISPHOSPHATE | | 分子名称: | ACETATE ION, CALCIUM ION, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, ... | | 著者 | Essen, L.-O, Perisic, O, Williams, R.L. | | 登録日 | 1996-08-24 | | 公開日 | 1997-07-07 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural mapping of the catalytic mechanism for a mammalian phosphoinositide-specific phospholipase C.
Biochemistry, 36, 1997
|
|
2EXV
 
 | | Crystal structure of the F7A mutant of the cytochrome c551 from Pseudomonas aeruginosa | | 分子名称: | ACETIC ACID, Cytochrome c-551, HEME C | | 著者 | Bonivento, D, Di Matteo, A, Borgia, A, Travaglini-Allocatelli, C, Brunori, M. | | 登録日 | 2005-11-09 | | 公開日 | 2006-02-07 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Unveiling a Hidden Folding Intermediate in c-Type Cytochromes by Protein Engineering
J.Biol.Chem., 281, 2006
|
|
1DF0
 
 | | Crystal structure of M-Calpain | | 分子名称: | CALPAIN, M-CALPAIN | | 著者 | Hosfield, C.M, Elce, J.S, Davies, P.L, Jia, Z. | | 登録日 | 1999-11-16 | | 公開日 | 2000-06-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of calpain reveals the structural basis for Ca(2+)-dependent protease activity and a novel mode of enzyme activation.
EMBO J., 18, 1999
|
|
7CE4
 
 | | Tankyrase2 catalytic domain in complex with K-476 | | 分子名称: | 5-[3-[[1-(6,7-dimethoxyquinazolin-4-yl)piperidin-4-yl]methyl]-2-oxidanylidene-4H-quinazolin-1-yl]-2-fluoranyl-benzenecarbonitrile, Poly [ADP-ribose] polymerase tankyrase-2, SULFATE ION, ... | | 著者 | Takahashi, Y, Suzuki, M, Saito, J. | | 登録日 | 2020-06-22 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The dual pocket binding novel tankyrase inhibitor K-476 enhances the efficacy of immune checkpoint inhibitor by attracting CD8 + T cells to tumors.
Am J Cancer Res, 11, 2021
|
|
5B5T
 
 | |
1IQW
 
 | | CRYSTAL STRUCTURE OF THE FAB FRAGMENT OF THE MOUSE ANTI-HUMAN FAS ANTIBODY HFE7A | | 分子名称: | ANTIBODY M-HFE7A, HEAVY CHAIN, LIGHT CHAIN | | 著者 | Ito, S, Takayama, T, Hanzawa, H, Ichikawa, K, Ohsumi, J, Serizawa, N, Hata, T, Haruyama, H. | | 登録日 | 2001-08-10 | | 公開日 | 2002-01-23 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of the antigen-binding fragment of apoptosis-inducing mouse anti-human Fas monoclonal antibody HFE7A.
J.Biochem., 131, 2002
|
|
5FLY
 
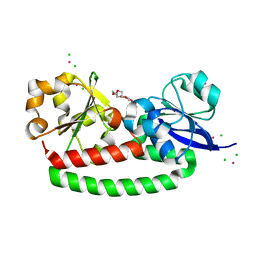 | | The FhuD protein from S.pseudintermedius | | 分子名称: | 2,5,8,11,14,17,20,23-OCTAOXAPENTACOSAN-25-OL, CADMIUM ION, CHLORIDE ION, ... | | 著者 | Malito, E. | | 登録日 | 2015-10-29 | | 公開日 | 2015-12-02 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.598 Å) | | 主引用文献 | Crystal Structure of Fhud at 1.6 Angstrom Resolution: A Ferrichrome-Binding Protein from the Animal and Human Pathogen Staphylococcus Pseudintermedius
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5GZ6
 
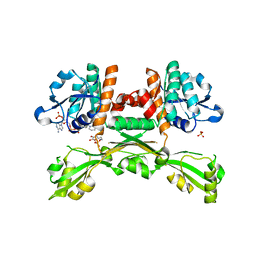 | | Structure of D-amino acid dehydrogenase in complex with NADPH and 2-keto-6-aminocapronic acid | | 分子名称: | 6-azanyl-2-oxidanylidene-hexanoic acid, ACETATE ION, Meso-diaminopimelate D-dehydrogenase, ... | | 著者 | Sakuraba, H, Seto, T, Hayashi, J, Akita, H, Yoneda, K, Ohshima, T. | | 登録日 | 2016-09-26 | | 公開日 | 2017-04-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Structure-Based Engineering of an Artificially Generated NADP+-Dependent d-Amino Acid Dehydrogenase
Appl. Environ. Microbiol., 83, 2017
|
|
5GZ3
 
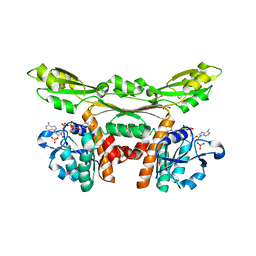 | | Structure of D-amino acid dehydrogenase in complex with NADP | | 分子名称: | 1,2-ETHANEDIOL, Meso-diaminopimelate D-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Sakuraba, H, Seto, T, Hayashi, J, Akita, H, Yoneda, K, Ohshima, T. | | 登録日 | 2016-09-26 | | 公開日 | 2017-04-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Structure-Based Engineering of an Artificially Generated NADP+-Dependent d-Amino Acid Dehydrogenase
Appl. Environ. Microbiol., 83, 2017
|
|
5GZ1
 
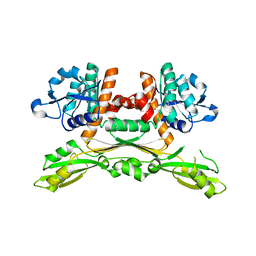 | | Structure of substrate/cofactor-free D-amino acid dehydrogenase | | 分子名称: | Meso-diaminopimelate D-dehydrogenase | | 著者 | Sakuraba, H, Seto, T, Hayashi, J, Akita, H, Yoneda, K, Ohshima, T. | | 登録日 | 2016-09-26 | | 公開日 | 2017-04-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Structure-Based Engineering of an Artificially Generated NADP+-Dependent d-Amino Acid Dehydrogenase
Appl. Environ. Microbiol., 83, 2017
|
|
5KZV
 
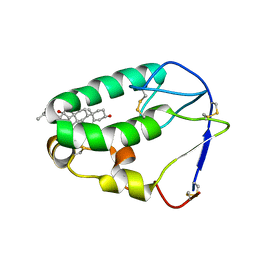 | | Crystal structure of the xenopus Smoothened cysteine-rich domain (CRD) in complex with 20(S)-hydroxycholesterol | | 分子名称: | (3alpha,8alpha)-cholest-5-ene-3,20-diol, Smoothened | | 著者 | Huang, P, Kim, Y, Salic, A. | | 登録日 | 2016-07-25 | | 公開日 | 2016-08-17 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.616 Å) | | 主引用文献 | Cellular Cholesterol Directly Activates Smoothened in Hedgehog Signaling.
Cell, 166, 2016
|
|
5KZY
 
 | |
5KZZ
 
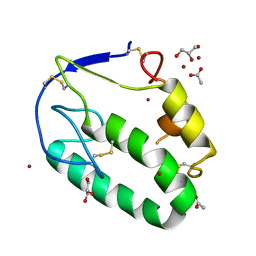 | | Crystal structure of the xenopus Smoothened cysteine-rich domain (CRD) in its apo-form | | 分子名称: | ACETATE ION, GLYCEROL, Smoothened, ... | | 著者 | Huang, P, Kim, Y, Salic, A. | | 登録日 | 2016-07-25 | | 公開日 | 2016-08-17 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.332 Å) | | 主引用文献 | Cellular Cholesterol Directly Activates Smoothened in Hedgehog Signaling.
Cell, 166, 2016
|
|
2E0O
 
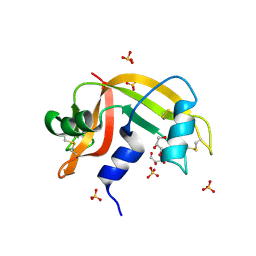 | | Mutant Human Ribonuclease 1 (V52L, D53L, N56L, F59L) | | 分子名称: | GLYCEROL, Ribonuclease, SULFATE ION | | 著者 | Yamada, H, Tamada, T, Kosaka, M, Kuroki, R. | | 登録日 | 2006-10-10 | | 公開日 | 2007-08-28 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | 'Crystal lattice engineering,' an approach to engineer protein crystal contacts by creating intermolecular symmetry: crystallization and structure determination of a mutant human RNase 1 with a hydrophobic interface of leucines
Protein Sci., 16, 2007
|
|
2E0L
 
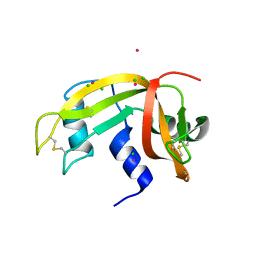 | | Mutant Human Ribonuclease 1 (Q28L, R31L, R32L) | | 分子名称: | CADMIUM ION, CHLORIDE ION, Ribonuclease | | 著者 | Yamada, H, Tamada, T, Kosaka, M, Kuroki, R. | | 登録日 | 2006-10-10 | | 公開日 | 2007-08-28 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | 'Crystal lattice engineering,' an approach to engineer protein crystal contacts by creating intermolecular symmetry: crystallization and structure determination of a mutant human RNase 1 with a hydrophobic interface of leucines
Protein Sci., 16, 2007
|
|
2E0J
 
 | | Mutant Human Ribonuclease 1 (R31L, R32L) | | 分子名称: | Ribonuclease | | 著者 | Yamada, H, Tamada, T, Kosaka, M, Kuroki, R. | | 登録日 | 2006-10-10 | | 公開日 | 2007-08-28 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | 'Crystal lattice engineering,' an approach to engineer protein crystal contacts by creating intermolecular symmetry: crystallization and structure determination of a mutant human RNase 1 with a hydrophobic interface of leucines
Protein Sci., 16, 2007
|
|
1NTD
 
 | | STRUCTURE OF ALCALIGENES FAECALIS NITRITE REDUCTASE MUTANT M150E THAT CONTAINS ZINC | | 分子名称: | COPPER (II) ION, NITRITE REDUCTASE | | 著者 | Murphy, M.E.P, Adman, E.T, Turley, S. | | 登録日 | 1995-07-03 | | 公開日 | 1996-11-08 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of Alcaligenes faecalis nitrite reductase and a copper site mutant, M150E, that contains zinc.
Biochemistry, 34, 1995
|
|
1VEC
 
 | | Crystal structure of the N-terminal domain of rck/p54, a human DEAD-box protein | | 分子名称: | ATP-dependent RNA helicase p54, L(+)-TARTARIC ACID, ZINC ION | | 著者 | Hogetsu, K, Matsui, T, Yukihiro, Y, Tanaka, M, Sato, T, Kumasaka, T, Tanaka, N. | | 登録日 | 2004-03-29 | | 公開日 | 2004-04-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Structural insight of human DEAD-box protein rck/p54 into its substrate recognition with conformational changes
Genes Cells, 11, 2006
|
|
