7DUP
 
 | | Apo structure of wild type Bt4394, a GH20 family sulfoglycosidase | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-N-acetylhexosaminidase, CHLORIDE ION, ... | | 著者 | Zhang, Z, He, Y, Jin, Y. | | 登録日 | 2021-01-10 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Mechanistic and Structural Insights into the Specificity and Biological Functions of Bacterial Sulfoglycosidases
Acs Catalysis, 13, 2023
|
|
7DVA
 
 | | Structure of wild type Bt4394, a GH20 family sulfoglycosidase, in complex with 6S-GlcNAc | | 分子名称: | 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, GLYCEROL | | 著者 | Zhang, Z, He, Y, Jin, Y. | | 登録日 | 2021-01-13 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Mechanistic and Structural Insights into the Specificity and Biological Functions of Bacterial Sulfoglycosidases
Acs Catalysis, 13, 2023
|
|
6AVS
 
 | | Complex structure of JMJD5 and Symmetric Monomethyl-Arginine (MMA) | | 分子名称: | (2S)-2-amino-5-[(N-methylcarbamimidoyl)amino]pentanoic acid, Lysine-specific demethylase 8, ZINC ION | | 著者 | Lee, S, Liu, H, Wang, Y, Dai, S, Zhang, G. | | 登録日 | 2017-09-04 | | 公開日 | 2018-02-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Specific Recognition of Arginine Methylated Histone Tails by JMJD5 and JMJD7.
Sci Rep, 8, 2018
|
|
7DVB
 
 | | D335N variant of Bt4394 in complex with 6SO3-NAG-oxazoline intermediate | | 分子名称: | 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, [(3~{a}~{R},5~{R},6~{S},7~{R},7~{a}~{R})-2-methyl-6,7-bis(oxidanyl)-5,6,7,7~{a}-tetrahydro-3~{a}~{H}-pyrano[3,2-d][1,3]oxazol-1-ium-5-yl]methyl sulfate | | 著者 | Zhang, Z, He, Y, Jin, Y. | | 登録日 | 2021-01-13 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Mechanistic and Structural Insights into the Specificity and Biological Functions of Bacterial Sulfoglycosidases
Acs Catalysis, 13, 2023
|
|
2JTC
 
 | | 3D structure and backbone dynamics of SPE B | | 分子名称: | Streptopain | | 著者 | Chuang, W, Wang, C, Houng, H, Chen, C, Wang, P. | | 登録日 | 2007-07-26 | | 公開日 | 2008-08-26 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure and backbone dynamics of streptopain: insight into diverse substrate specificity.
J.Biol.Chem., 284, 2009
|
|
6AX3
 
 | | Complex structure of JMJD5 and Symmetric Dimethyl-Arginine (SDMA) | | 分子名称: | 2-OXOGLUTARIC ACID, Lysine-specific demethylase 8, N3, ... | | 著者 | Lee, S, Liu, H, Wang, Y, Dai, S, Zhang, G. | | 登録日 | 2017-09-06 | | 公開日 | 2018-02-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Specific Recognition of Arginine Methylated Histone Tails by JMJD5 and JMJD7.
Sci Rep, 8, 2018
|
|
7XJG
 
 | | Cryo-EM structure of E.coli retron-Ec86 in complex with its effector at 2.5 angstrom | | 分子名称: | DNA (105-MER), MAGNESIUM ION, RNA (14-MER), ... | | 著者 | Wang, Y.J, Guan, Z.Y, Zou, T.T. | | 登録日 | 2022-04-17 | | 公開日 | 2022-09-14 | | 最終更新日 | 2025-06-25 | | 実験手法 | ELECTRON MICROSCOPY (2.51 Å) | | 主引用文献 | Cryo-EM structures of Escherichia coli Ec86 retron complexes reveal architecture and defence mechanism.
Nat Microbiol, 7, 2022
|
|
6WEQ
 
 | | DENV1 NS1 in complex with neutralizing 2B7 Fab fragment | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2B7 Fab fragment heavy chain, 2B7 Fab fragment light chain, ... | | 著者 | Akey, D.L, Smith, J.L. | | 登録日 | 2020-04-02 | | 公開日 | 2020-12-23 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural basis for antibody inhibition of flavivirus NS1-triggered endothelial dysfunction.
Science, 371, 2021
|
|
4I0R
 
 | | Crystal structure of spleen tyrosine kinase complexed with 2-(3,4,5-Trimethoxy-phenyl)-5H-pyrrolo[2,3-b]pyrazine-7-carboxylic acid isopropylamide | | 分子名称: | N-(propan-2-yl)-2-(3,4,5-trimethoxyphenyl)-5H-pyrrolo[2,3-b]pyrazine-7-carboxamide, Tyrosine-protein kinase SYK | | 著者 | Kuglstatter, A, Villasenor, A.G. | | 登録日 | 2012-11-19 | | 公開日 | 2013-10-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Pyrrolopyrazines as selective spleen tyrosine kinase inhibitors.
J.Med.Chem., 56, 2013
|
|
6LXT
 
 | | Structure of post fusion core of 2019-nCoV S2 subunit | | 分子名称: | Spike protein S2, TETRAETHYLENE GLYCOL, ZINC ION | | 著者 | Zhu, Y, Sun, F. | | 登録日 | 2020-02-11 | | 公開日 | 2020-02-26 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion.
Cell Res., 30, 2020
|
|
4I0T
 
 | | Crystal structure of spleen tyrosine kinase complexed with 2-(5,6,7,8-Tetrahydro-imidazo[1,5-a]pyridin-1-yl)-5H-pyrrolo[2,3-b]pyrazine-7-carboxylic acid tert-butylamide | | 分子名称: | N-tert-butyl-2-(5,6,7,8-tetrahydroimidazo[1,5-a]pyridin-1-yl)-5H-pyrrolo[2,3-b]pyrazine-7-carboxamide, Tyrosine-protein kinase SYK | | 著者 | Kuglstatter, A, Slade, M. | | 登録日 | 2012-11-19 | | 公開日 | 2013-10-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Pyrrolopyrazines as selective spleen tyrosine kinase inhibitors.
J.Med.Chem., 56, 2013
|
|
9GC6
 
 | |
9GC5
 
 | |
9GC4
 
 | |
9GDV
 
 | |
7VZG
 
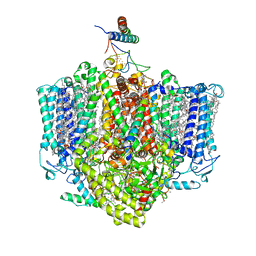 | | Structure of the Acidobacteria homodimeric reaction center bound with cytochrome c (the larger form) | | 分子名称: | BACTERIOCHLOROPHYLL A, CALCIUM ION, CHLOROPHYLL A, ... | | 著者 | Huang, G.Q, Dong, S.S, Qin, X.C, Sui, S.F. | | 登録日 | 2021-11-16 | | 公開日 | 2023-02-22 | | 実験手法 | ELECTRON MICROSCOPY (2.61 Å) | | 主引用文献 | Structure of the Acidobacteria homodimeric reaction center bound with cytochrome c.
Nat Commun, 13, 2022
|
|
5J3F
 
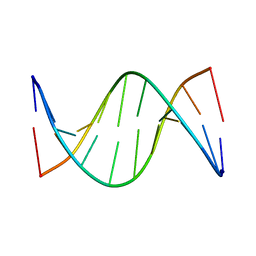 | | NMR solution structure of [Rp, Rp]-PT dsDNA | | 分子名称: | DNA (5'-D(*CP*GP*(RSG)P*CP*CP*GP*CP*CP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*GP*CP*GP*(RSG)P*CP*CP*G)-3') | | 著者 | Lan, W, Hu, Z, Cao, C. | | 登録日 | 2016-03-30 | | 公開日 | 2016-11-16 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural investigation into physiological DNA phosphorothioate modification
Sci Rep, 6, 2016
|
|
2H5E
 
 | |
5J3G
 
 | |
4I0S
 
 | |
2HJD
 
 | |
7RZU
 
 | |
7RZS
 
 | |
7RZR
 
 | |
8WJH
 
 | | Cryo-EM structure of OAT4 | | 分子名称: | CHLORIDE ION, Solute carrier family 22 member 11 | | 著者 | Qian, H.W, He, J.J. | | 登録日 | 2023-09-26 | | 公開日 | 2024-08-28 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis for the transport and substrate selection of human urate transporter 1.
Cell Rep, 43, 2024
|
|
