7CL0
 
 | | Crystal structure of human SIRT6 | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NAD-dependent protein deacetylase sirtuin-6, ... | | 著者 | Song, K, Zhang, J. | | 登録日 | 2020-07-20 | | 公開日 | 2021-02-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.53 Å) | | 主引用文献 | Reply to: Binding site for MDL-801 on SIRT6.
Nat.Chem.Biol., 17, 2021
|
|
7CL1
 
 | |
7VK9
 
 | | Crystal structure of xCas9 P411T | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1 | | 著者 | Bao, R, Liu, H.Y, Luo, Y.Z, Song, Y.J. | | 登録日 | 2021-09-29 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural and Dynamics Studies of the Spcas9 Variant Provide Insights into the Regulatory Role of the REC1 Domain
Acs Catalysis, 12, 2022
|
|
2JVA
 
 | | NMR solution structure of peptidyl-tRNA hydrolase domain protein from Pseudomonas syringae pv. tomato. Northeast Structural Genomics Consortium target PsR211 | | 分子名称: | Peptidyl-tRNA hydrolase domain protein | | 著者 | Singarapu, K.K, Sukumaran, D, Parish, D, Eletsky, A, Zhang, Q, Zhao, L, Jiang, M, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Huang, Y.J, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2007-09-14 | | 公開日 | 2007-10-02 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of the peptidyl-tRNA hydrolase domain from Pseudomonas syringae expands the structural coverage of the hydrolysis domains of class 1 peptide chain release factors.
Proteins, 71, 2008
|
|
2L1V
 
 | |
7CMK
 
 | | E30 E-particle in complex with 6C5 | | 分子名称: | Heavy chain, Light chain, VP1, ... | | 著者 | Wang, K, Zhu, F, Rao, Z, Wang, X. | | 登録日 | 2020-07-27 | | 公開日 | 2020-08-12 | | 最終更新日 | 2020-09-16 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Serotype specific epitopes identified by neutralizing antibodies underpin immunogenic differences in Enterovirus B.
Nat Commun, 11, 2020
|
|
7W5S
 
 | | A nonheme iron- and alpha-ketoglutarate- dependent halogenase that catalyzes nucleotide substrates | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FE (II) ION, ... | | 著者 | Dai, L.H, Zhang, X, Hu, Y.M, Huang, J.W, Chen, C.C, Guo, R.T. | | 登録日 | 2021-11-30 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Structural and Functional Insights into a Nonheme Iron- and alpha-Ketoglutarate-Dependent Halogenase That Catalyzes Chlorination of Nucleotide Substrates.
Appl.Environ.Microbiol., 88, 2022
|
|
7W5T
 
 | | A nonheme iron- and alpha-ketoglutarate- dependent halogenase that catalyzes nucleotide substrates | | 分子名称: | 2-OXOGLUTARIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Dai, L.H, Zhang, X, Hu, Y.M, Huang, J.W, Chen, C.C, Guo, R.T. | | 登録日 | 2021-11-30 | | 公開日 | 2022-04-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Structural and Functional Insights into a Nonheme Iron- and alpha-Ketoglutarate-Dependent Halogenase That Catalyzes Chlorination of Nucleotide Substrates.
Appl.Environ.Microbiol., 88, 2022
|
|
7W5V
 
 | | A nonheme iron- and alpha-ketoglutarate- dependent halogenase that catalyzes nucleotide substrates | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FE (III) ION, ... | | 著者 | Dai, L.H, Zhang, X, Hu, Y.M, Huang, J.W, Chen, C.C, Guo, R.T. | | 登録日 | 2021-11-30 | | 公開日 | 2022-04-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Structural and Functional Insights into a Nonheme Iron- and alpha-Ketoglutarate-Dependent Halogenase That Catalyzes Chlorination of Nucleotide Substrates.
Appl.Environ.Microbiol., 88, 2022
|
|
5WRV
 
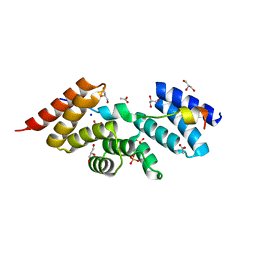 | | Complex structure of human SRP72/SRP68 | | 分子名称: | ACETATE ION, GLYCEROL, SODIUM ION, ... | | 著者 | Gao, Y, Chen, Z. | | 登録日 | 2016-12-04 | | 公開日 | 2017-06-21 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Human apo-SRP72 and SRP68/72 complex structures reveal the molecular basis of protein translocation
J Mol Cell Biol, 9, 2017
|
|
5WBG
 
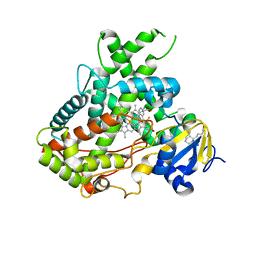 | | Crystal Structure of human Cytochrome P450 2B6 (Y226H/K262R) in complex with an analog of a drug Efavirenz | | 分子名称: | (2R,4S)-6-chloro-4-(cyclopropylethynyl)-2-methyl-4-(trifluoromethyl)-1,4-dihydro-2H-3,1-benzoxazine, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, 5-cyclohexylpentan-1-ol, ... | | 著者 | Shah, M.B, Halpert, J.R. | | 登録日 | 2017-06-29 | | 公開日 | 2018-05-02 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.99 Å) | | 主引用文献 | Crystal Structure of CYP2B6 in Complex with an Efavirenz Analog.
Int J Mol Sci, 19, 2018
|
|
5WQN
 
 | |
5WQO
 
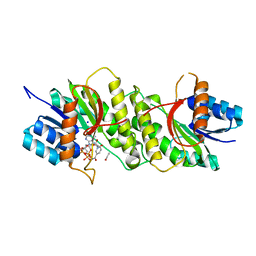 | | Crystal structure of a carbonyl reductase from Pseudomonas aeruginosa PAO1 in complex with NADP (condition I) | | 分子名称: | 1,2-ETHANEDIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Probable dehydrogenase, ... | | 著者 | Li, S, Wang, Y, Bartlam, M. | | 登録日 | 2016-11-27 | | 公開日 | 2017-10-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Structure and characterization of a NAD(P)H-dependent carbonyl reductase from Pseudomonas aeruginosa PAO1.
FEBS Lett., 591, 2017
|
|
5WQM
 
 | |
5X23
 
 | | Crystal structure of CYP2C9 genetic variant A477T (*30) in complex with multiple losartan molecules | | 分子名称: | Cytochrome P450 2C9, PHOSPHATE ION, POTASSIUM ION, ... | | 著者 | Maekawa, K, Adachi, M, Shah, M.B. | | 登録日 | 2017-01-30 | | 公開日 | 2017-10-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Basis of Single-Nucleotide Polymorphisms in Cytochrome P450 2C9
Biochemistry, 56, 2017
|
|
5X24
 
 | | Crystal structure of CYP2C9 genetic variant I359L (*3) in complex with multiple losartan molecules | | 分子名称: | Cytochrome P450 2C9, PHOSPHATE ION, POTASSIUM ION, ... | | 著者 | Maekawa, K, Adachi, M, Shah, M.B. | | 登録日 | 2017-01-30 | | 公開日 | 2017-10-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Structural Basis of Single-Nucleotide Polymorphisms in Cytochrome P450 2C9
Biochemistry, 56, 2017
|
|
5WQP
 
 | | Crystal structure of a carbonyl reductase from Pseudomonas aeruginosa PAO1 in complex with NADP (condition II) | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NICOTINAMIDE, PHOSPHATE ION, ... | | 著者 | Li, S, Wang, Y, Bartlam, M. | | 登録日 | 2016-11-27 | | 公開日 | 2017-10-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure and characterization of a NAD(P)H-dependent carbonyl reductase from Pseudomonas aeruginosa PAO1.
FEBS Lett., 591, 2017
|
|
5WRW
 
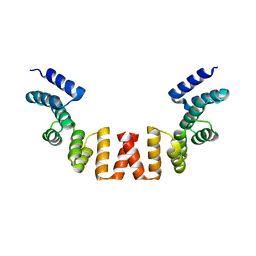 | | Structure of human apo-SRP72 | | 分子名称: | SULFATE ION, Signal recognition particle subunit SRP72 | | 著者 | Gao, Y, Chen, Z. | | 登録日 | 2016-12-04 | | 公開日 | 2017-06-21 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.91 Å) | | 主引用文献 | Human apo-SRP72 and SRP68/72 complex structures reveal the molecular basis of protein translocation
J Mol Cell Biol, 9, 2017
|
|
8GRR
 
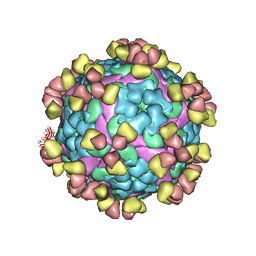 | |
8GSP
 
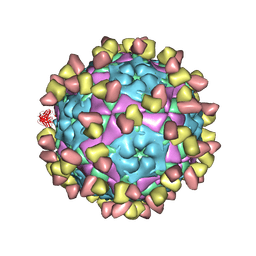 | | Complex of FMDV A/WH/CHA/09 and bovine neutralizing scFv antibody W2 | | 分子名称: | A/WH/CHA/09 VP1, A/WH/CHA/09 VP2, A/WH/CHA/09 VP3, ... | | 著者 | He, Y, Li, K. | | 登録日 | 2022-09-06 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-12-06 | | 実験手法 | ELECTRON MICROSCOPY (3.75 Å) | | 主引用文献 | Conserved antigen structures and antibody-driven variations on foot-and-mouth disease virus serotype A revealed by bovine neutralizing monoclonal antibodies.
Plos Pathog., 19, 2023
|
|
5XVA
 
 | | Crystal Structure of PAK4 in complex with inhibitor CZH216 | | 分子名称: | ETHANOL, Serine/threonine-protein kinase PAK 4, [6-chloranyl-4-[(5-methyl-1H-pyrazol-3-yl)amino]quinazolin-2-yl]-[(3R)-3-methylpiperazin-1-yl]methanone | | 著者 | Zhao, F, Li, H. | | 登録日 | 2017-06-27 | | 公開日 | 2018-02-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.847 Å) | | 主引用文献 | Structure-Based Design of 6-Chloro-4-aminoquinazoline-2-carboxamide Derivatives as Potent and Selective p21-Activated Kinase 4 (PAK4) Inhibitors.
J. Med. Chem., 61, 2018
|
|
5XVG
 
 | | Crystal Structure of PAK4 in complex with inhibitor CZH226 | | 分子名称: | 1,2-ETHANEDIOL, ETHANOL, Serine/threonine-protein kinase PAK 4, ... | | 著者 | Zhao, F, Li, H. | | 登録日 | 2017-06-27 | | 公開日 | 2018-02-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure-Based Design of 6-Chloro-4-aminoquinazoline-2-carboxamide Derivatives as Potent and Selective p21-Activated Kinase 4 (PAK4) Inhibitors.
J. Med. Chem., 61, 2018
|
|
8HJB
 
 | |
5XA6
 
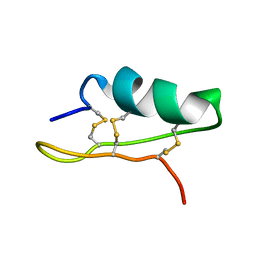 | |
8HC0
 
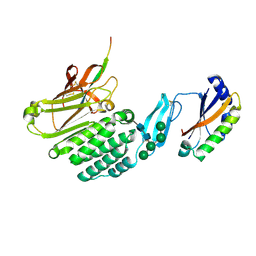 | | Crystal structure of the extracellular domains of GPR110 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Adhesion G-protein coupled receptor F1, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Wang, F.F, Song, G.J. | | 登録日 | 2022-11-01 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal Structure of the Extracellular Domains of GPR110.
J.Mol.Biol., 435, 2023
|
|
