2BRB
 
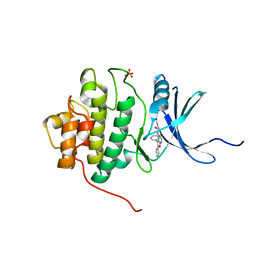 | | Structure-based Design of Novel Chk1 Inhibitors: Insights into Hydrogen Bonding and Protein-Ligand Affinity | | 分子名称: | 2-[(5,6-DIPHENYLFURO[2,3-D]PYRIMIDIN-4-YL)AMINO]ETHANOL, SERINE/THREONINE-PROTEIN KINASE CHK1, SULFATE ION | | 著者 | Foloppe, N, Fisher, L.M, Howes, R, Kierstan, P, Potter, A, Robertson, A.G.S, Surgenor, A.E. | | 登録日 | 2005-05-04 | | 公開日 | 2005-05-12 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure-Based Design of Novel Chk1 Inhibitors: Insights Into Hydrogen Bonding and Protein-Ligand Affinity.
J.Med.Chem., 48, 2005
|
|
6XKK
 
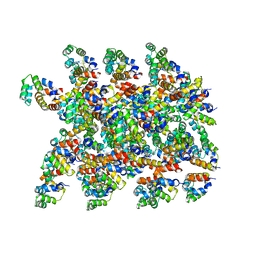 | | Cryo-EM structure of the NLRP1-CARD filament | | 分子名称: | NACHT, LRR and PYD domains-containing protein 1 | | 著者 | Hollingsworth, L.R, David, L, Li, Y, Sharif, H, Fontana, P, Fu, T, Wu, H. | | 登録日 | 2020-06-26 | | 公開日 | 2020-11-25 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.72 Å) | | 主引用文献 | Mechanism of filament formation in UPA-promoted CARD8 and NLRP1 inflammasomes.
Nat Commun, 12, 2021
|
|
1MOT
 
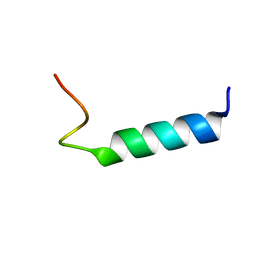 | | NMR Structure Of Extended Second Transmembrane Domain Of Glycine Receptor alpha1 Subunit in SDS Micelles | | 分子名称: | Glycine Receptor alpha-1 CHAIN | | 著者 | Yushmanov, V.E, Mandal, P.K, Liu, Z, Tang, P, Xu, Y. | | 登録日 | 2002-09-09 | | 公開日 | 2003-09-23 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR Structure and Backbone Dynamics of the Extended Second Transmembrane Domain of the Human Neuronal Glycine Receptor Alpha1 Subunit
Biochemistry, 42, 2003
|
|
6X6C
 
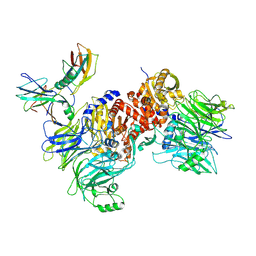 | | Cryo-EM structure of NLRP1-DPP9-VbP complex | | 分子名称: | Dipeptidyl peptidase 9, NACHT, LRR and PYD domains-containing protein 1, ... | | 著者 | Hollingsworth, L.R, Sharif, H, Griswold, A.R, Fontana, P, Mintseris, J, Dagbay, K.B, Paulo, J.A, Gygi, S.P, Bachovchin, D.A, Wu, H. | | 登録日 | 2020-05-27 | | 公開日 | 2021-03-10 | | 最終更新日 | 2021-05-12 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | DPP9 sequesters the C terminus of NLRP1 to repress inflammasome activation.
Nature, 592, 2021
|
|
6X6A
 
 | | Cryo-EM structure of NLRP1-DPP9 complex | | 分子名称: | Dipeptidyl peptidase 9, NACHT, LRR and PYD domains-containing protein 1 | | 著者 | Hollingsworth, L.R, Sharif, H, Griswold, A.R, Fontana, P, Mintseris, J, Dagbay, K.B, Paulo, J.A, Gygi, S.P, Bachovchin, D.A, Wu, H. | | 登録日 | 2020-05-27 | | 公開日 | 2021-03-10 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | DPP9 sequesters the C terminus of NLRP1 to repress inflammasome activation.
Nature, 592, 2021
|
|
6XKJ
 
 | | Cryo-EM structure of CARD8-CARD filament | | 分子名称: | Caspase recruitment domain-containing protein 8 | | 著者 | Hollingsworth, L.R, David, L, Li, Y, Sharif, H, Fontana, P, Fu, T, Wu, H. | | 登録日 | 2020-06-26 | | 公開日 | 2020-11-25 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.54 Å) | | 主引用文献 | Mechanism of filament formation in UPA-promoted CARD8 and NLRP1 inflammasomes.
Nat Commun, 12, 2021
|
|
4Z90
 
 | | ELIC bound with the anesthetic isoflurane in the resting state | | 分子名称: | (2R)-2-chloro-2-(difluoromethoxy)-1,1,1-trifluoroethane, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Gamma-aminobutyric-acid receptor subunit beta-1 | | 著者 | Chen, Q, Kinde, M, Arjunan, P, Cohen, A, Xu, Y, Tang, P. | | 登録日 | 2015-04-09 | | 公開日 | 2015-09-16 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Direct Pore Binding as a Mechanism for Isoflurane Inhibition of the Pentameric Ligand-gated Ion Channel ELIC.
Sci Rep, 5, 2015
|
|
3R5L
 
 | | Structure of Ddn, the Deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Deazaflavin-dependent nitroreductase | | 著者 | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayyar, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | 登録日 | 2011-03-18 | | 公開日 | 2012-01-18 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3R5R
 
 | | Structure of Ddn, the Deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824, with co-factor F420 | | 分子名称: | COENZYME F420, Deazaflavin-dependent nitroreductase | | 著者 | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | 登録日 | 2011-03-19 | | 公開日 | 2012-01-18 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.101 Å) | | 主引用文献 | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
4Z91
 
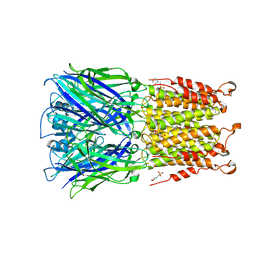 | | ELIC cocrystallized with isofluorane in a desensitized state | | 分子名称: | (2R)-2-chloro-2-(difluoromethoxy)-1,1,1-trifluoroethane, 1.7.6 3-bromanylpropan-1-amine, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | 著者 | Chen, Q, Kinde, M.N, Arjunan, P, Cohen, A, Xu, Y, Tang, P. | | 登録日 | 2015-04-09 | | 公開日 | 2015-09-16 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.3915 Å) | | 主引用文献 | Direct Pore Binding as a Mechanism for Isoflurane Inhibition of the Pentameric Ligand-gated Ion Channel ELIC.
Sci Rep, 5, 2015
|
|
3R5Z
 
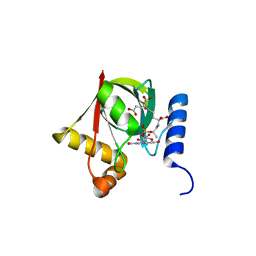 | | Structure of a Deazaflavin-dependent reductase from Nocardia farcinica, with co-factor F420 | | 分子名称: | COENZYME F420, Putative uncharacterized protein, SULFATE ION | | 著者 | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | 登録日 | 2011-03-20 | | 公開日 | 2012-01-18 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.503 Å) | | 主引用文献 | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3R5P
 
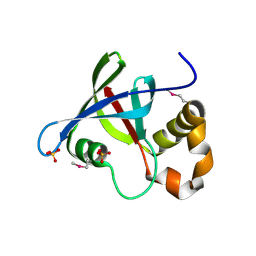 | | Structure of Ddn, the Deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824 | | 分子名称: | Deazaflavin-dependent nitroreductase, SULFATE ION | | 著者 | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | 登録日 | 2011-03-19 | | 公開日 | 2012-01-18 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3R5Y
 
 | | Structure of a Deazaflavin-dependent nitroreductase from Nocardia farcinica, with co-factor F420 | | 分子名称: | COENZYME F420, Putative uncharacterized protein | | 著者 | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | 登録日 | 2011-03-20 | | 公開日 | 2012-01-18 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.801 Å) | | 主引用文献 | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3R5W
 
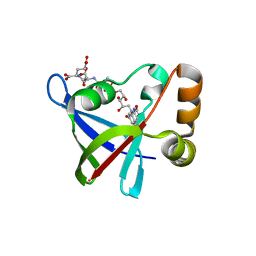 | | Structure of Ddn, the Deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824, with co-factor F420 | | 分子名称: | COENZYME F420, Deazaflavin-dependent nitroreductase | | 著者 | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | 登録日 | 2011-03-20 | | 公開日 | 2012-01-18 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.786 Å) | | 主引用文献 | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3QQ5
 
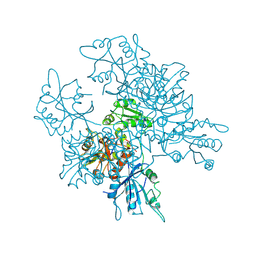 | | Crystal structure of the [FeFe]-hydrogenase maturation protein HydF | | 分子名称: | Small GTP-binding protein | | 著者 | Cendron, L, Berto, P, D'Adamo, S, Vallese, F, Govoni, C, Posewitz, M.C, Giacometti, G.M, Costantini, P, Zanotti, G. | | 登録日 | 2011-02-15 | | 公開日 | 2011-11-16 | | 最終更新日 | 2012-01-11 | | 実験手法 | X-RAY DIFFRACTION (2.99 Å) | | 主引用文献 | Crystal Structure of HydF Scaffold Protein Provides Insights into [FeFe]-Hydrogenase Maturation.
J.Biol.Chem., 286, 2011
|
|
7RPM
 
 | |
2V7A
 
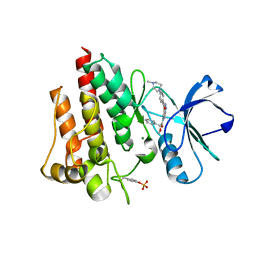 | | Crystal structure of the T315I Abl mutant in complex with the inhibitor PHA-739358 | | 分子名称: | MAGNESIUM ION, N-[(3E)-5-[(2R)-2-METHOXY-2-PHENYLACETYL]PYRROLO[3,4-C]PYRAZOL-3(5H)-YLIDENE]-4-(4-METHYLPIPERAZIN-1-YL)BENZAMIDE, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE ABL1 | | 著者 | Modugno, M, Casale, E, Soncini, C, Rosettani, P, Colombo, R, Lupi, R, Rusconi, L, Fancelli, D, Carpinelli, P, Cameron, A.D, Isacchi, A, Moll, J. | | 登録日 | 2007-07-27 | | 公開日 | 2007-09-18 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structure of the T315I Abl Mutant in Complex with the Aurora Kinases Inhibitor Pha-739358.
Cancer Res., 67, 2007
|
|
2V79
 
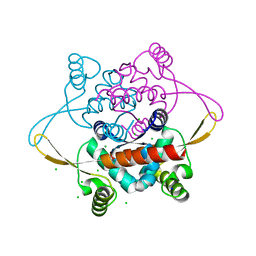 | | Crystal Structure of the N-terminal domain of DnaD from Bacillus Subtilis | | 分子名称: | CHLORIDE ION, DNA REPLICATION PROTEIN DNAD, SODIUM ION | | 著者 | Schneider, S, Zhang, W, Soultanas, P, Paoli, M. | | 登録日 | 2007-07-27 | | 公開日 | 2008-01-15 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of the N-Terminal Oligomerization Domain of Dnad Reveals a Unique Tetramerization Motif and Provides Insights Into Scaffold Formation.
J.Mol.Biol., 376, 2008
|
|
5SXU
 
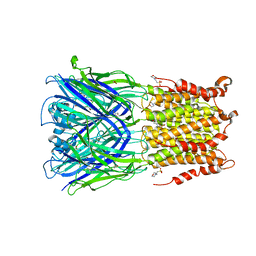 | | X-ray structure of 2-bromoethanol bound to a pentameric ligand gated ion channel (ELIC) in a desensitized state | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-BROMOETHANOL, 3-AMINOPROPANE, ... | | 著者 | Chen, Q, Kinde, M, Cohen, A, Xu, Y, Tang, P. | | 登録日 | 2016-08-10 | | 公開日 | 2017-06-21 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural Basis of Alcohol Inhibition of the Pentameric Ligand-Gated Ion Channel ELIC.
Structure, 25, 2017
|
|
5SXV
 
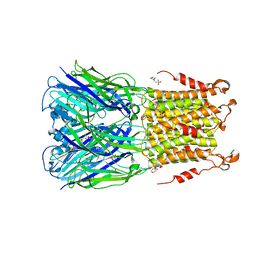 | | X-ray structure of 2-bromoethanol bound to a pentameric ligand gated ion channel (ELIC) in a resting state | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-BROMOETHANOL, Cys-loop ligand-gated ion channel | | 著者 | Chen, Q, Kinde, M, Cohen, A, Xu, Y, Tang, P. | | 登録日 | 2016-08-10 | | 公開日 | 2017-06-21 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Structural Basis of Alcohol Inhibition of the Pentameric Ligand-Gated Ion Channel ELIC.
Structure, 25, 2017
|
|
2M6I
 
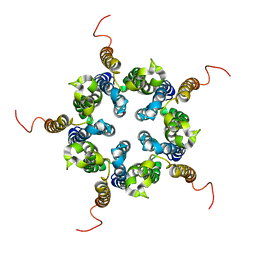 | | Putative pentameric open-channel structure of full-length transmembrane domains of human glycine receptor alpha1 subunit | | 分子名称: | Full-Length Transmembrane Domains of Human Glycine Receptor alpha1 Subunit | | 著者 | Mowrey, D, Cui, T, Jia, Y, Ma, D, Makhov, A.M, Zhang, P, Tang, P, Xu, Y. | | 登録日 | 2013-03-29 | | 公開日 | 2013-09-04 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Open-Channel Structures of the Human Glycine Receptor alpha 1 Full-Length Transmembrane Domain.
Structure, 21, 2013
|
|
3PJR
 
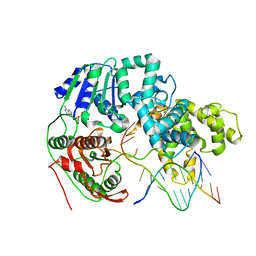 | | HELICASE SUBSTRATE COMPLEX | | 分子名称: | 5'-D(*CP*GP*AP*GP*CP*AP*CP*TP*GP*C)-3', 5'-D(*GP*CP*AP*GP*TP*GP*CP*TP*CP*GP*TP*TP*TP*TP*T)-3', ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Velankar, S.S, Soultanas, P, Dillingham, M.S, Subramanya, H.S, Wigley, D.B. | | 登録日 | 1999-03-12 | | 公開日 | 1999-04-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism
Cell(Cambridge,Mass.), 97, 1999
|
|
2M6B
 
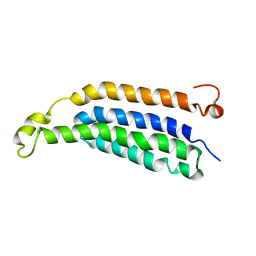 | | Structure of full-length transmembrane domains of human glycine receptor alpha1 monomer subunit | | 分子名称: | Full-Length Transmembrane Domains of Human Glycine Receptor alpha1 Subunit | | 著者 | Mowrey, D, Cui, T, Jia, Y, Ma, D, Makhov, A.M, Zhang, P, Tang, P, Xu, Y. | | 登録日 | 2013-03-28 | | 公開日 | 2013-09-04 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Open-Channel Structures of the Human Glycine Receptor alpha 1 Full-Length Transmembrane Domain.
Structure, 21, 2013
|
|
4F8H
 
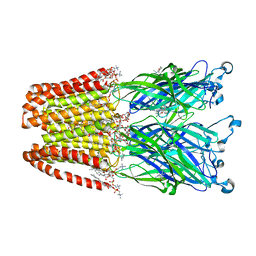 | | X-ray Structure of the Anesthetic Ketamine Bound to the GLIC Pentameric Ligand-gated Ion Channel | | 分子名称: | (R)-ketamine, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Proton-gated ion channel, ... | | 著者 | Pan, J.J, Chen, Q, Willenbring, D, Kong, X.P, Cohen, A, Xu, Y, Tang, P. | | 登録日 | 2012-05-17 | | 公開日 | 2012-08-29 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.99 Å) | | 主引用文献 | Structure of the Pentameric Ligand-Gated Ion Channel GLIC Bound with Anesthetic Ketamine.
Structure, 20, 2012
|
|
3RQW
 
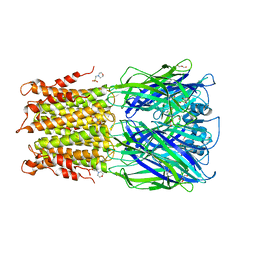 | | Crystal structure of acetylcholine bound to a prokaryotic pentameric ligand-gated ion channel, ELIC | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETYLCHOLINE, ELIC Pentameric Ligand Gated Ion Channel from Erwinia Chrysanthemi, ... | | 著者 | Pan, J.J, Chen, Q, Yoshida, K, Cohen, A, Kong, X.P, Xu, Y, Tang, P. | | 登録日 | 2011-04-28 | | 公開日 | 2012-03-07 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.913 Å) | | 主引用文献 | Structure of the pentameric ligand-gated ion channel ELIC cocrystallized with its competitive antagonist acetylcholine.
Nat Commun, 3, 2012
|
|
