6LFW
 
 | |
6LCS
 
 | | Crystal structure of 73MuL9 Fv-clasp fragment in complex with GA-pyridine analogue | | 分子名称: | (2~{S})-6-[4-(hydroxymethyl)-3-oxidanyl-pyridin-1-ium-1-yl]-2-(phenylmethoxycarbonylamino)hexanoic acid, PHOSPHATE ION, VH-SARAH, ... | | 著者 | Nakamura, T, Takagi, J, Yamagata, Y, Morioka, H. | | 登録日 | 2019-11-19 | | 公開日 | 2020-11-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Molecular recognition of a single-chain Fv antibody specific for GA-pyridine, an advanced glycation end-product (AGE), elucidated using biophysical techniques and synthetic antigen analogues.
J.Biochem., 170, 2021
|
|
6LFV
 
 | |
5X3Z
 
 | | Solution structure of musashi1 RBD2 in complex with RNA | | 分子名称: | RNA (5'-R(*GP*UP*AP*GP*U)-3'), RNA-binding protein Musashi homolog 1 | | 著者 | Iwaoka, R, Nagata, T, Tsuda, K, Imai, T, Okano, H, Kobayashi, N, Katahira, M. | | 登録日 | 2017-02-09 | | 公開日 | 2017-12-13 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Insight into the Recognition of r(UAG) by Musashi-1 RBD2, and Construction of a Model of Musashi-1 RBD1-2 Bound to the Minimum Target RNA
Molecules, 22, 2017
|
|
5X3Y
 
 | | Refined solution structure of musashi1 RBD2 | | 分子名称: | RNA-binding protein Musashi homolog 1 | | 著者 | Iwaoka, R, Nagata, T, Tsuda, K, Imai, T, Okano, H, Kobayashi, N, Katahira, M. | | 登録日 | 2017-02-09 | | 公開日 | 2017-12-13 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Insight into the Recognition of r(UAG) by Musashi-1 RBD2, and Construction of a Model of Musashi-1 RBD1-2 Bound to the Minimum Target RNA
Molecules, 22, 2017
|
|
1ITY
 
 | | Solution structure of the DNA binding domain of human TRF1 | | 分子名称: | TRF1 | | 著者 | Nishikawa, T, Okamura, H, Nagadoi, A, Konig, P, Rhodes, D, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2002-02-15 | | 公開日 | 2002-03-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a telomeric DNA complex of human TRF1
Structure, 9, 2001
|
|
1IV6
 
 | | Solution Structure of the DNA Complex of Human TRF1 | | 分子名称: | 5'-D(*CP*CP*CP*TP*AP*AP*CP*CP*CP*TP*AP*AP*C)-3', 5'-D(*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*G)-3', TELOMERIC REPEAT BINDING FACTOR 1 | | 著者 | Nishikawa, T, Okamura, H, Nagadoi, A, Konig, P, Rhodes, D, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2002-03-14 | | 公開日 | 2002-04-17 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a telomeric DNA complex of human TRF1.
Structure, 9, 2001
|
|
2LQI
 
 | | NMR structure of FOXO3a transactivation domains (CR2C-CR3) in complex with CBP KIX domain (2l3b conformation) | | 分子名称: | CREB-binding protein, Forkhead box O3 | | 著者 | Wang, F, Marshall, C.B, Yamamoto, K, Li, G.B, Gasmi-Seabrook, G.M.C, Okada, H, Mak, T.W, Ikura, M. | | 登録日 | 2012-03-06 | | 公開日 | 2012-05-16 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structures of KIX domain of CBP in complex with two FOXO3a transactivation domains reveal promiscuity and plasticity in coactivator recruitment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2ZWS
 
 | | Crystal Structure Analysis of neutral ceramidase from Pseudomonas aeruginosa | | 分子名称: | FORMIC ACID, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Kakuta, Y, Okino, N, Inoue, T, Okano, H, Ito, M. | | 登録日 | 2008-12-17 | | 公開日 | 2009-03-03 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Mechanistic insights into the hydrolysis and synthesis of ceramide by neutral ceramidase.
J.Biol.Chem., 284, 2009
|
|
2Z33
 
 | | Solution structure of the DNA complex of PhoB DNA-binding/transactivation Domain | | 分子名称: | 5'-D(*AP*CP*AP*GP*AP*TP*TP*TP*AP*TP*GP*AP*CP*AP*GP*T)-3', 5'-D(*AP*CP*TP*GP*TP*CP*AP*TP*AP*AP*AP*TP*CP*TP*GP*T)-3', Phosphate regulon transcriptional regulatory protein phoB | | 著者 | Yamane, T, Okamura, H, Ikeguchi, M, Nishimura, Y, Kidera, A. | | 登録日 | 2007-05-31 | | 公開日 | 2008-04-22 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Water-mediated interactions between DNA and PhoB DNA-binding/transactivation domain: NMR-restrained molecular dynamics in explicit water environment.
Proteins, 71, 2008
|
|
2E1A
 
 | | crystal structure of FFRP-DM1 | | 分子名称: | 75aa long hypothetical regulatory protein AsnC, SELENOMETHIONINE | | 著者 | Koike, H, Suzuki, M. | | 登録日 | 2006-10-19 | | 公開日 | 2007-09-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A Structural Code for Discriminating between Transcription Signals Revealed by the Feast/Famine Regulatory Protein DM1 in Complex with Ligands
Structure, 15, 2007
|
|
2Z4P
 
 | | Crystal structure of FFRP-DM1 | | 分子名称: | 75aa long hypothetical regulatory protein AsnC, ISOLEUCINE | | 著者 | Yamada, M, Koike, H, Kudo, N, Suzuki, M. | | 登録日 | 2007-06-21 | | 公開日 | 2007-09-18 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | A Structural Code for Discriminating between Transcription Signals Revealed by the Feast/Famine Regulatory Protein DM1 in Complex with Ligands
Structure, 15, 2007
|
|
2LX4
 
 | |
4J20
 
 | | X-ray structure of the cytochrome c-554 from chlorobaculum tepidum | | 分子名称: | Cytochrome c-555, HEME B/C, ISOPROPYL ALCOHOL, ... | | 著者 | Unno, M, Yu, L.J, Wang-otomo, Z.Y. | | 登録日 | 2013-02-04 | | 公開日 | 2013-10-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Structure analysis and characterization of the cytochrome c-554 from thermophilic green sulfur photosynthetic bacterium Chlorobaculum tepidum
Photosynth.Res., 118, 2013
|
|
1X1D
 
 | | Crystal structure of BchU complexed with S-adenosyl-L-homocysteine and Zn-bacteriopheophorbide d | | 分子名称: | CrtF-related protein, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | 著者 | Yamaguchi, H, Wada, K, Fukuyama, K. | | 登録日 | 2005-04-04 | | 公開日 | 2006-07-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal Structures of BchU, a Methyltransferase Involved in Bacteriochlorophyll c Biosynthesis, and its Complex with S-adenosylhomocysteine: Implications for Reaction Mechanism.
J.Mol.Biol., 360, 2006
|
|
1X1B
 
 | | Crystal structure of BchU complexed with S-adenosyl-L-homocysteine | | 分子名称: | CrtF-related protein, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | 著者 | Yamaguchi, H, Wada, K, Fukuyama, K. | | 登録日 | 2005-04-03 | | 公開日 | 2006-07-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal Structures of BchU, a Methyltransferase Involved in Bacteriochlorophyll c Biosynthesis, and its Complex with S-adenosylhomocysteine: Implications for Reaction Mechanism.
J.Mol.Biol., 360, 2006
|
|
1X19
 
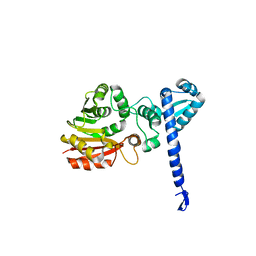 | | Crystal structure of BchU involved in bacteriochlorophyll c biosynthesis | | 分子名称: | CrtF-related protein, SULFATE ION | | 著者 | Yamaguchi, H, Wada, K, Fukuyama, K. | | 登録日 | 2005-04-02 | | 公開日 | 2006-07-18 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Crystal Structures of BchU, a Methyltransferase Involved in Bacteriochlorophyll c Biosynthesis, and its Complex with S-adenosylhomocysteine: Implications for Reaction Mechanism.
J.Mol.Biol., 360, 2006
|
|
1X1C
 
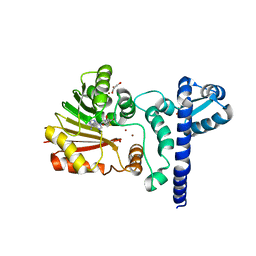 | | Crystal structure of BchU complexed with S-adenosyl-L-homocysteine and Zn2+ | | 分子名称: | CrtF-related protein, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | 著者 | Yamaguchi, H, Wada, K, Fukuyama, K. | | 登録日 | 2005-04-03 | | 公開日 | 2006-07-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Crystal Structures of BchU, a Methyltransferase Involved in Bacteriochlorophyll c Biosynthesis, and its Complex with S-adenosylhomocysteine: Implications for Reaction Mechanism.
J.Mol.Biol., 360, 2006
|
|
1X1A
 
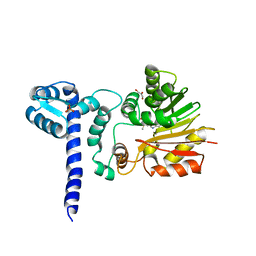 | | Crystal structure of BchU complexed with S-adenosyl-L-methionine | | 分子名称: | CrtF-related protein, GLYCEROL, S-ADENOSYLMETHIONINE, ... | | 著者 | Yamaguchi, H, Wada, K, Fukuyama, K. | | 登録日 | 2005-04-03 | | 公開日 | 2006-07-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal Structures of BchU, a Methyltransferase Involved in Bacteriochlorophyll c Biosynthesis, and its Complex with S-adenosylhomocysteine: Implications for Reaction Mechanism.
J.Mol.Biol., 360, 2006
|
|
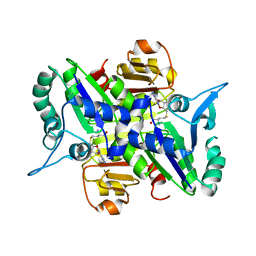
2E0N
 
 | |
2E0K
 
 | |
2YQH
 
 | | Crystal structure of uridine-diphospho-N-acetylglucosamine pyrophosphorylase from Candida albicans, in the substrate-binding form | | 分子名称: | 2-acetamido-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Miki, K, Maruyama, D, Nishitani, Y, Nonaka, T, Kita, A. | | 登録日 | 2007-03-30 | | 公開日 | 2007-05-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal Structure of Uridine-diphospho-N-acetylglucosamine Pyrophosphorylase from Candida albicans and Catalytic Reaction Mechanism
J.Biol.Chem., 282, 2007
|
|
8ILL
 
 | | Crystal structure of a highly photostable and bright green fluorescent protein at pH5.6 | | 分子名称: | CHLORIDE ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, green fluorescent protein | | 著者 | Ago, H, Ando, R, Hirano, M, Shimozono, S, Miyawaki, A, Yamamoto, M. | | 登録日 | 2023-03-03 | | 公開日 | 2023-10-04 | | 最終更新日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | StayGold variants for molecular fusion and membrane-targeting applications.
Nat.Methods, 21, 2024
|
|
8ILK
 
 | | Crystal structure of a highly photostable and bright green fluorescent protein at pH8.5 | | 分子名称: | CHLORIDE ION, Green FLUORESCENT PROTEIN | | 著者 | Ago, H, Ando, R, Hirano, M, Shimozono, S, Miyawaki, A, Yamamoto, M. | | 登録日 | 2023-03-03 | | 公開日 | 2023-10-04 | | 最終更新日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | StayGold variants for molecular fusion and membrane-targeting applications.
Nat.Methods, 21, 2024
|
|
5NEN
 
 | |
