5GUA
 
 | | Structure of biotin carboxyl carrier protein from pyrococcus horikoshi OT3 (delta N79) A138Y mutant | | 分子名称: | 149aa long hypothetical methylmalonyl-CoA decarboxylase gamma chain | | 著者 | Yamada, K, Kunishima, N, Matsuura, Y, Nakai, K, Naitow, H, Fukasawa, Y, Tomii, K. | | 登録日 | 2016-08-26 | | 公開日 | 2017-08-30 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Designing better diffracting crystals of biotin carboxyl carrier protein from Pyrococcus horikoshii by a mutation based on the crystal-packing propensity of amino acids.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5GU9
 
 | | Structure of biotin carboxyl carrier protein from pyrococcus horikoshi OT3 (delta N79) A138I mutant | | 分子名称: | 149aa long hypothetical methylmalonyl-CoA decarboxylase gamma chain | | 著者 | Yamada, K, Kunishima, N, Matsuura, Y, Nakai, K, Naitow, H, Fukasawa, Y, Tomii, K. | | 登録日 | 2016-08-26 | | 公開日 | 2017-08-30 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Designing better diffracting crystals of biotin carboxyl carrier protein from Pyrococcus horikoshii by a mutation based on the crystal-packing propensity of amino acids.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5GU8
 
 | | Structure of biotin carboxyl carrier protein from pyrococcus horikoshi OT3 (delta N79) wild type | | 分子名称: | 149aa long hypothetical methylmalonyl-CoA decarboxylase gamma chain, SODIUM ION | | 著者 | Yamada, K, Kunishima, N, Matsuura, Y, Nakai, K, Naitow, H, Fukasawa, Y, Tomii, K. | | 登録日 | 2016-08-26 | | 公開日 | 2017-08-30 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Designing better diffracting crystals of biotin carboxyl carrier protein from Pyrococcus horikoshii by a mutation based on the crystal-packing propensity of amino acids.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
7VPY
 
 | | Crystal structure of the neutralizing nanobody P86 against SARS-CoV-2 | | 分子名称: | 1,2-ETHANEDIOL, Nanobody, SULFATE ION | | 著者 | Maeda, R, Fujita, J, Konishi, Y, Kazuma, Y, Yamazaki, H, Anzai, I, Yamaguchi, K, Kasai, K, Nagata, K, Yamaoka, Y, Miyakawa, K, Ryo, A, Shirakawa, K, Makino, F, Matsuura, Y, Inoue, T, Imura, A, Namba, K, Takaori-Kondo, A. | | 登録日 | 2021-10-18 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including Omicron.
Commun Biol, 5, 2022
|
|
2CDV
 
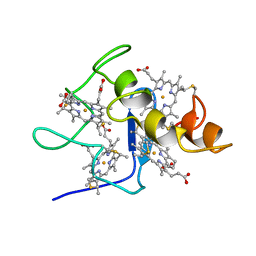 | | REFINED STRUCTURE OF CYTOCHROME C3 AT 1.8 ANGSTROMS RESOLUTION | | 分子名称: | CYTOCHROME C3, HEME C | | 著者 | Higuchi, Y, Kusunoki, M, Matsuura, Y, Yasuoka, N, Kakudo, M. | | 登録日 | 1983-11-15 | | 公開日 | 1984-02-02 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Refined structure of cytochrome c3 at 1.8 A resolution
J.Mol.Biol., 172, 1984
|
|
7VQ0
 
 | | Cryo-EM structure of the SARS-CoV-2 spike protein (2-up RBD) bound to neutralizing nanobodies P86 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Maeda, R, Fujita, J, Konishi, Y, Kazuma, Y, Yamazaki, H, Anzai, I, Yamaguchi, K, Kasai, K, Nagata, K, Yamaoka, Y, Miyakawa, K, Ryo, A, Shirakawa, K, Makino, F, Matsuura, Y, Inoue, T, Imura, A, Namba, K, Takaori-Kondo, A. | | 登録日 | 2021-10-18 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.03 Å) | | 主引用文献 | A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including Omicron.
Commun Biol, 5, 2022
|
|
1IV8
 
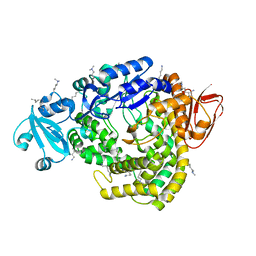 | |
5H2X
 
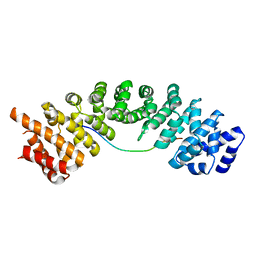 | |
5H2W
 
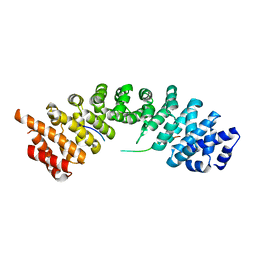 | |
5H2V
 
 | |
2BFH
 
 | | CRYSTAL STRUCTURE OF BASIC FIBROBLAST GROWTH FACTOR AT 1.6 ANGSTROMS RESOLUTION | | 分子名称: | BASIC FIBROBLAST GROWTH FACTOR | | 著者 | Kitagawa, Y, Ago, H, Katsube, Y, Fujishima, A, Matsuura, Y. | | 登録日 | 1993-08-31 | | 公開日 | 1994-01-31 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of basic fibroblast growth factor at 1.6 A resolution.
J.Biochem.(Tokyo), 110, 1991
|
|
2EL2
 
 | | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (L185M) | | 分子名称: | S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, diphthine synthase | | 著者 | Asada, Y, Tanaka, Y, Pampa, K, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-03-26 | | 公開日 | 2007-10-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (L185M)
To be Published
|
|
2AMG
 
 | | STRUCTURE OF HYDROLASE (GLYCOSIDASE) | | 分子名称: | 1,4-ALPHA-D-GLUCAN MALTOTETRAHYDROLASE, CALCIUM ION | | 著者 | Morishita, Y, Hasegawa, K, Matsuura, Y, Kubota, M, Sakai, S, Katsube, Y. | | 登録日 | 1996-12-23 | | 公開日 | 1997-04-01 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of a maltotetraose-forming exo-amylase from Pseudomonas stutzeri.
J.Mol.Biol., 267, 1997
|
|
2EK2
 
 | | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (E140M) | | 分子名称: | S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, diphthine synthase | | 著者 | Asada, Y, Matsuura, Y, Tanaka, Y, Nakamoto, T, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-03-22 | | 公開日 | 2007-09-25 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (E140M)
To be Published
|
|
2ZTN
 
 | | Hepatitis E virus ORF2 (Genotype 3) | | 分子名称: | Capsid protein | | 著者 | Yamashita, T, Unno, H, Mori, Y, Li, T.C, Takeda, N, Matsuura, Y. | | 登録日 | 2008-10-08 | | 公開日 | 2009-08-25 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.56 Å) | | 主引用文献 | Biological and immunological characteristics of hepatitis E virus-like particles based on the crystal structure
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2EK7
 
 | | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (L163M) | | 分子名称: | Probable diphthine synthase, S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION | | 著者 | Asada, Y, Taketa, M, Morikawa, Y, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-03-22 | | 公開日 | 2007-09-25 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (L163M)
To be Published
|
|
2EKB
 
 | | Structural study of Project ID TTHB049 from Thermus thermophilus HB8 (L19M) | | 分子名称: | Alpha-ribazole-5'-phosphate phosphatase, SODIUM ION | | 著者 | Asada, Y, Taketa, M, Tanaka, Y, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-03-22 | | 公開日 | 2007-09-25 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural study of Project ID TTHB049 from Thermus thermophilus HB8 (L19M)
To be Published
|
|
2EKA
 
 | | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (L202M) | | 分子名称: | S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, diphthine synthase | | 著者 | Asada, Y, Matsuura, Y, Kageyama, Y, Nakamoto, T, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-03-22 | | 公開日 | 2007-09-25 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (L202M)
To be Published
|
|
5XZX
 
 | |
5ZGO
 
 | |
5XW5
 
 | |
5X8N
 
 | |
5XOJ
 
 | | Crystal structure of Xpo1p-PKI-Nup42p-Gsp1p-GTP complex | | 分子名称: | Exportin-1, GTP-binding nuclear protein, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Koyama, M, Shirai, N, Matsuura, Y. | | 登録日 | 2017-05-29 | | 公開日 | 2017-08-16 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of the Xpo1p nuclear export complex bound to the SxFG/PxFG repeats of the nucleoporin Nup42p
Genes Cells, 22, 2017
|
|
5XW4
 
 | |
5XGS
 
 | |
