7CUI
 
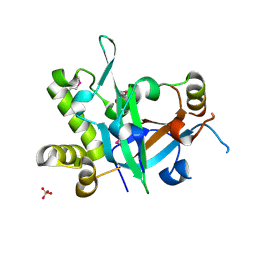 | | Crystal structure of fission yeast Pot1 and Tpz1 | | 分子名称: | Protection of telomeres protein 1, Protection of telomeres protein tpz1, SULFATE ION | | 著者 | Sun, H, Wu, Z, Wu, J, Lei, M. | | 登録日 | 2020-08-23 | | 公開日 | 2021-08-25 | | 最終更新日 | 2022-09-07 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural insights into Pot1-ssDNA, Pot1-Tpz1 and Tpz1-Ccq1 Interactions within fission yeast shelterin complex.
Plos Genet., 18, 2022
|
|
5VLR
 
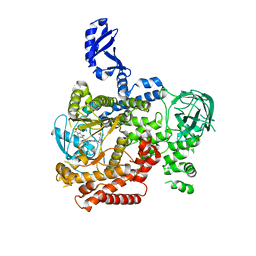 | | CRYSTAL STRUCTURE OF PI3K DELTA IN COMPLEX WITH A TRIFLUORO-ETHYL-PYRAZOL-PYROLOTRIAZINE INHIBITOR | | 分子名称: | 4-acetyl-1-(3-{4-amino-5-[1-(2,2,2-trifluoroethyl)-1H-pyrazol-5-yl]pyrrolo[2,1-f][1,2,4]triazin-7-yl}phenyl)-3,3-dimethylpiperazin-2-one, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | 著者 | Sack, J.S. | | 登録日 | 2017-04-26 | | 公開日 | 2017-06-07 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Identification of a Potent, Selective, and Efficacious Phosphatidylinositol 3-Kinase delta (PI3K delta ) Inhibitor for the Treatment of Immunological Disorders.
J. Med. Chem., 60, 2017
|
|
4ALN
 
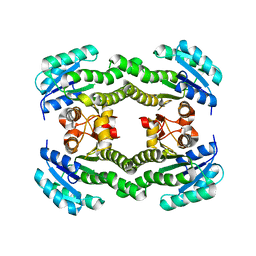 | | Crystal structure of S. aureus FabI (P32) | | 分子名称: | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH] | | 著者 | Schiebel, J, Kisker, C. | | 登録日 | 2012-03-04 | | 公開日 | 2012-05-09 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | Staphylococcus Aureus Fabi: Inhibition, Substrate Recognition and Potential Implications for in Vivo Essentiality
Structure, 20, 2012
|
|
4ALI
 
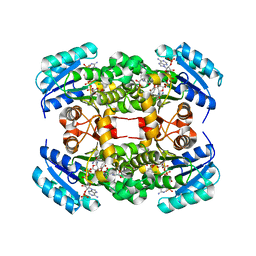 | | Crystal structure of S. aureus FabI in complex with NADP and triclosan (P1) | | 分子名称: | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], GLUTAMIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Schiebel, J, Chang, A, Tonge, P.J, Kisker, C. | | 登録日 | 2012-03-04 | | 公開日 | 2012-05-09 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Staphylococcus Aureus Fabi: Inhibition, Substrate Recognition and Potential Implications for in Vivo Essentiality
Structure, 20, 2012
|
|
4ALK
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 5-ethyl- 2-phenoxyphenol | | 分子名称: | 5-ETHYL-2-PHENOXYPHENOL, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], GLUTAMIC ACID, ... | | 著者 | Schiebel, J, Chang, A, Tonge, P.J, Kisker, C. | | 登録日 | 2012-03-04 | | 公開日 | 2012-05-09 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Staphylococcus Aureus Fabi: Inhibition, Substrate Recognition and Potential Implications for in Vivo Essentiality
Structure, 20, 2012
|
|
4ALL
 
 | | Crystal structure of S. aureus FabI in complex with NADP and triclosan (P212121) | | 分子名称: | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TRICLOSAN | | 著者 | Schiebel, J, Chang, A, Tonge, P.J, Kisker, C. | | 登録日 | 2012-03-04 | | 公開日 | 2012-05-09 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Staphylococcus Aureus Fabi: Inhibition, Substrate Recognition and Potential Implications for in Vivo Essentiality
Structure, 20, 2012
|
|
4ALJ
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 5-chloro- 2-phenoxyphenol | | 分子名称: | 5-CHLORO-2-PHENOXYPHENOL, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], GLUTAMIC ACID, ... | | 著者 | Schiebel, J, Chang, A, Tonge, P.J, Kisker, C. | | 登録日 | 2012-03-04 | | 公開日 | 2012-05-09 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Staphylococcus Aureus Fabi: Inhibition, Substrate Recognition and Potential Implications for in Vivo Essentiality
Structure, 20, 2012
|
|
4ALM
 
 | | Crystal structure of S. aureus FabI (P43212) | | 分子名称: | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], SULFATE ION | | 著者 | Schiebel, J, Kisker, C. | | 登録日 | 2012-03-04 | | 公開日 | 2012-05-09 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Staphylococcus Aureus Fabi: Inhibition, Substrate Recognition and Potential Implications for in Vivo Essentiality
Structure, 20, 2012
|
|
3ZU5
 
 | | Structure of the enoyl-ACP reductase FabV from Yersinia pestis with the cofactor NADH and the 2-pyridone inhibitor PT173 | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 1-(3-amino-2-methylbenzyl)-4-hexylpyridin-2(1H)-one, PUTATIVE REDUCTASE YPO4104/Y4119/YP_4011, ... | | 著者 | Hirschbeck, M.W, Kuper, J, Tonge, P.J, Kisker, C. | | 登録日 | 2011-07-13 | | 公開日 | 2012-01-18 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of the Yersinia Pestis Fabv Enoyl-Acp Reductase and its Interaction with Two 2-Pyridone Inhibitors
Structure, 20, 2012
|
|
3ZU3
 
 | | Structure of the enoyl-ACP reductase FabV from Yersinia pestis with the cofactor NADH (MR, cleaved Histag) | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, PUTATIVE REDUCTASE YPO4104/Y4119/YP_4011, ... | | 著者 | Hirschbeck, M.W, Kuper, J, Kisker, C. | | 登録日 | 2011-07-13 | | 公開日 | 2012-01-18 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.802 Å) | | 主引用文献 | Structure of the Yersinia Pestis Fabv Enoyl-Acp Reductase and its Interaction with Two 2-Pyridone Inhibitors
Structure, 20, 2012
|
|
3ZU2
 
 | | Structure of the enoyl-ACP reductase FabV from Yersinia pestis with the cofactor NADH (SIRAS) | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, PUTATIVE REDUCTASE YPO4104/Y4119/YP_4011, SODIUM ION | | 著者 | Hirschbeck, M.W, Kuper, J, Kisker, C. | | 登録日 | 2011-07-13 | | 公開日 | 2012-01-18 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of the Yersinia Pestis Fabv Enoyl-Acp Reductase and its Interaction with Two 2-Pyridone Inhibitors
Structure, 20, 2012
|
|
3ZU4
 
 | | Structure of the enoyl-ACP reductase FabV from Yersinia pestis with the cofactor NADH and the 2-pyridone inhibitor PT172 | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 1-(2-CHLOROBENZYL)-4-HEXYLPYRIDIN-2(1H)-ONE, PUTATIVE REDUCTASE YPO4104/Y4119/YP_4011, ... | | 著者 | Hirschbeck, M.W, Kuper, J, Tonge, P.J, Kisker, C. | | 登録日 | 2011-07-13 | | 公開日 | 2012-01-18 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Structure of the Yersinia Pestis Fabv Enoyl-Acp Reductase and its Interaction with Two 2-Pyridone Inhibitors
Structure, 20, 2012
|
|
8D1Y
 
 | |
8D1P
 
 | | Crystal structure of Plasmodium falciparum GRP78-NBD in complex with 7-Deaza-2'-C-methyladenosine | | 分子名称: | 7-(2-C-methyl-beta-D-ribofuranosyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Chaperone DnaK, SULFATE ION | | 著者 | Mrozek, A, Chen, Y, Antoshchenko, T, Park, H.W. | | 登録日 | 2022-05-27 | | 公開日 | 2023-05-31 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | A non-traditional crystal-based compound screening method targeting the ATP binding site of Plasmodium falciparum GRP78 for identification of novel nucleoside analogues.
Front Mol Biosci, 9, 2022
|
|
8D24
 
 | | Crystal structure of Plasmodium falciparum GRP78-NBD in complex with VER155008 | | 分子名称: | 4-[[(2R,3S,4R,5R)-5-[6-amino-8-[(3,4-dichlorophenyl)methylamino]purin-9-yl]-3,4-dihydroxy-oxolan-2-yl]methoxymethyl]benzonitrile, Chaperone DnaK, SULFATE ION | | 著者 | Mrozek, A, Park, H.W. | | 登録日 | 2022-05-27 | | 公開日 | 2023-05-31 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | A non-traditional crystal-based compound screening method targeting the ATP binding site of Plasmodium falciparum GRP78 for identification of novel nucleoside analogues.
Front Mol Biosci, 9, 2022
|
|
8D20
 
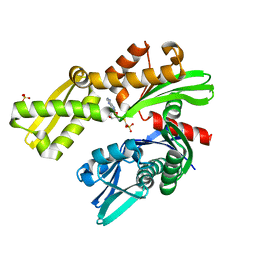 | |
8D1S
 
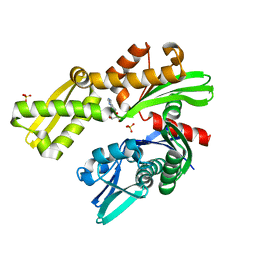 | | Crystal structure of Plasmodium falciparum GRP78 in complex with Toyocamycin | | 分子名称: | 4-amino-7-(beta-D-ribofuranosyl)-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Chaperone DnaK, SULFATE ION | | 著者 | Mrozek, A, Chen, Y, Antoshchenko, T, Park, H.W. | | 登録日 | 2022-05-27 | | 公開日 | 2023-05-31 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | A non-traditional crystal-based compound screening method targeting the ATP binding site of Plasmodium falciparum GRP78 for identification of novel nucleoside analogues.
Front Mol Biosci, 9, 2022
|
|
8D1Q
 
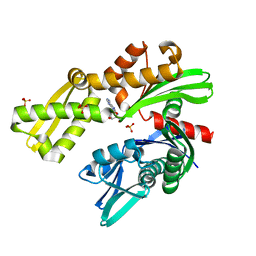 | | Crystal structure of Plasmodium falciparum GRP78-NBD in complex with 8-Aminoadenosine | | 分子名称: | (2R,3R,4S,5R)-2-(6,8-diaminopurin-9-yl)-5-(hydroxymethyl)oxolane-3,4-diol, Chaperone DnaK, SULFATE ION | | 著者 | Mrozek, A, Chen, Y, Antoshchenko, T, Park, H.W. | | 登録日 | 2022-05-27 | | 公開日 | 2023-05-31 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | A non-traditional crystal-based compound screening method targeting the ATP binding site of Plasmodium falciparum GRP78 for identification of novel nucleoside analogues.
Front Mol Biosci, 9, 2022
|
|
8D22
 
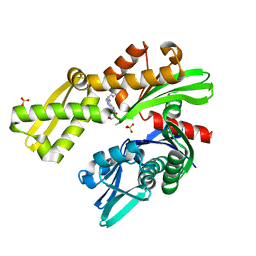 | |
5L1Z
 
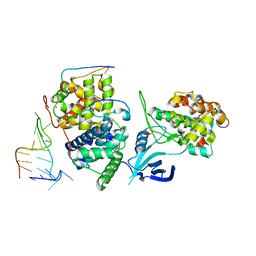 | | TAR complex with HIV-1 Tat-AFF4-P-TEFb | | 分子名称: | AF4/FMR2 family member 4, Cyclin-T1, Cyclin-dependent kinase 9, ... | | 著者 | Schulze-Gahmen, U, Hurley, J. | | 登録日 | 2016-07-29 | | 公開日 | 2016-10-26 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (5.9 Å) | | 主引用文献 | Insights into HIV-1 proviral transcription from integrative structure and dynamics of the Tat:AFF4:P-TEFb:TAR complex.
Elife, 5, 2016
|
|
6VY2
 
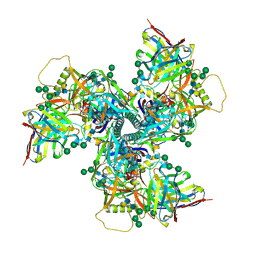 | |
6VU2
 
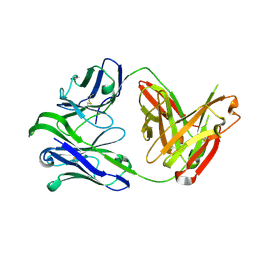 | | M1214_N1 Fab structure | | 分子名称: | M1214 N1 Fab heavy chain, M1214 N1 Fab light chain | | 著者 | Pan, R, Kong, X. | | 登録日 | 2020-02-14 | | 公開日 | 2020-05-06 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | VSV-Displayed HIV-1 Envelope Identifies Broadly Neutralizing Antibodies Class-Switched to IgG and IgA.
Cell Host Microbe, 27, 2020
|
|
8W9W
 
 | | The cryo-EM structure of human sphingomyelin synthase-related protein in complex with ceramide/phosphoethanolamine | | 分子名称: | PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER, Sphingomyelin synthase-related protein 1, ~{N}-[(~{Z},2~{S},3~{R})-1,3-bis(oxidanyl)heptadec-4-en-2-yl]dodecanamide | | 著者 | Hu, K, Zhang, Q, Chen, Y, Yao, D, Zhou, L, Cao, Y. | | 登録日 | 2023-09-06 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.74 Å) | | 主引用文献 | Cryo-EM structure of human sphingomyelin synthase and its mechanistic implications for sphingomyelin synthesis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8W9Y
 
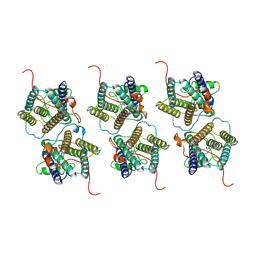 | | The cryo-EM structure of human sphingomyelin synthase-related protein | | 分子名称: | Sphingomyelin synthase-related protein 1 | | 著者 | Hu, K, Zhang, Q, Chen, Y, Yao, D, Zhou, L, Cao, Y. | | 登録日 | 2023-09-06 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Cryo-EM structure of human sphingomyelin synthase and its mechanistic implications for sphingomyelin synthesis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6LRD
 
 | |
