5C5Q
 
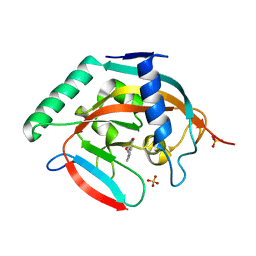 | | CRYSTAL STRUCTURE OF HUMAN TANKYRASE-2 IN COMPLEX WITH A PYRANOPYRIDONE INHIBITOR | | 分子名称: | (3R)-10-methyl-3-(propan-2-yl)-1,3,4,5-tetrahydro-6H-pyrano[4,3-c]isoquinolin-6-one, SULFATE ION, Tankyrase-2, ... | | 著者 | Lukacs, C.M, Janson, C.A. | | 登録日 | 2015-06-21 | | 公開日 | 2015-08-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Fragment-Based Drug Design of Novel Pyranopyridones as Cell Active and Orally Bioavailable Tankyrase Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
5HZG
 
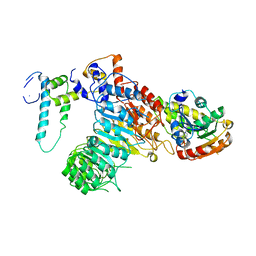 | | The crystal structure of the strigolactone-induced AtD14-D3-ASK1 complex | | 分子名称: | (2Z)-2-methylbut-2-ene-1,4-diol, F-box/LRR-repeat MAX2 homolog, SKP1-like protein 1A, ... | | 著者 | Yao, R.F, Ming, Z.H, Yan, L.M, Rao, Z.H, Lou, Z.Y, Xie, D.X. | | 登録日 | 2016-02-02 | | 公開日 | 2016-08-03 | | 最終更新日 | 2016-08-31 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | DWARF14 is a non-canonical hormone receptor for strigolactone
Nature, 536, 2016
|
|
5HYW
 
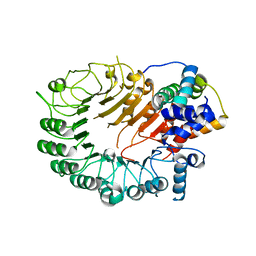 | | The crystal structure of the D3-ASK1 complex | | 分子名称: | F-box/LRR-repeat MAX2 homolog, SKP1-like protein 1A | | 著者 | Yao, R.F, Ming, Z.H, Yan, L.M, Rao, Z.H, Lou, Z.Y, Xie, D.X. | | 登録日 | 2016-02-02 | | 公開日 | 2016-08-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.01 Å) | | 主引用文献 | DWARF14 is a non-canonical hormone receptor for strigolactone
Nature, 536, 2016
|
|
8SIT
 
 | |
8SIS
 
 | |
8SIR
 
 | |
8SIQ
 
 | |
8SWH
 
 | |
6LQD
 
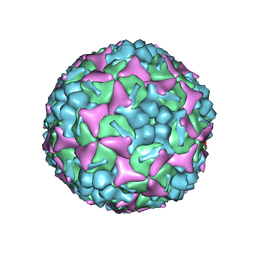 | | Structure of Enterovirus 71 in complex with NLD-22 | | 分子名称: | 1-(2-azanylpyridin-4-yl)-3-[5-[4-(5-methyl-1,2,4-oxadiazol-3-yl)phenoxy]pentyl]imidazolidin-2-one, Capsid protein VP1, Capsid protein VP2, ... | | 著者 | Zhang, M, Sun, Y, Wang, X, Guo, Y, Rao, Z. | | 登録日 | 2020-01-13 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.264 Å) | | 主引用文献 | Design, Synthesis, and Evaluation of Novel Enterovirus 71 Inhibitors as Therapeutic Drug Leads for the Treatment of Human Hand, Foot, and Mouth Disease.
J.Med.Chem., 63, 2020
|
|
8IMW
 
 | |
7DQZ
 
 | | Crystal structure of SARS 3C-like protease in apo form | | 分子名称: | 3C-like proteinase | | 著者 | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | 登録日 | 2020-12-24 | | 公開日 | 2021-09-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
7EO7
 
 | | Crystal structure of HCoV-NL63 3C-like protease in complex with an inhibitor Shikonin | | 分子名称: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | 著者 | Gao, H.X, Zhang, Y.T, Zhou, X.L, Zhong, F.L, Li, J, Zhang, J. | | 登録日 | 2021-04-21 | | 公開日 | 2021-10-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.24916625 Å) | | 主引用文献 | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
7EO8
 
 | | Crystal structure of SARS coronavirus main protease in complex with an inhibitor Shikonin | | 分子名称: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | 著者 | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | 登録日 | 2021-04-21 | | 公開日 | 2021-10-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.2808516 Å) | | 主引用文献 | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
6KVN
 
 | |
8H3X
 
 | |
8H3Y
 
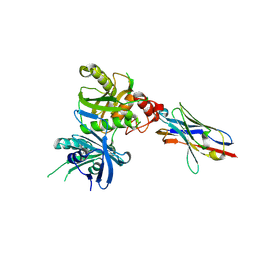 | |
6LCT
 
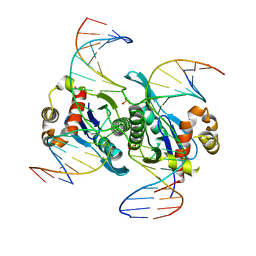 | | Crystal structure of catalytic inactive chloroplast resolvase NtMOC1 in complex with Holliday junction | | 分子名称: | DNA (5'-D(*AP*AP*GP*AP*TP*GP*TP*CP*CP*CP*TP*CP*TP*GP*TP*TP*GP*T)-3'), DNA (5'-D(*AP*CP*AP*AP*CP*AP*GP*AP*GP*GP*AP*TP*GP*GP*AP*GP*CP*T)-3'), DNA (5'-D(*GP*CP*CP*TP*TP*GP*CP*TP*GP*GP*GP*AP*CP*AP*TP*CP*TP*T)-3'), ... | | 著者 | Yan, J.J, Hong, S.X, Guan, Z.Y, Yin, P. | | 登録日 | 2019-11-19 | | 公開日 | 2020-04-08 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structural insights into sequence-dependent Holliday junction resolution by the chloroplast resolvase MOC1.
Nat Commun, 11, 2020
|
|
6KVO
 
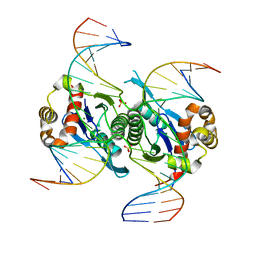 | | Crystal structure of chloroplast resolvase in complex with Holliday junction | | 分子名称: | DNA (5'-D(*AP*CP*AP*AP*CP*AP*GP*AP*TP*GP*AP*TP*GP*GP*AP*GP*CP*T)-3'), DNA (5'-D(*GP*CP*CP*TP*TP*GP*CP*TP*TP*GP*GP*AP*CP*AP*TP*CP*TP*T)-3'), DNA (5'-D(P*AP*AP*GP*AP*TP*GP*TP*CP*CP*AP*TP*CP*TP*GP*TP*TP*GP*T)-3'), ... | | 著者 | Yan, J.J, Hong, S.X, Guan, Z.Y, Yin, P. | | 登録日 | 2019-09-05 | | 公開日 | 2020-04-08 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural insights into sequence-dependent Holliday junction resolution by the chloroplast resolvase MOC1.
Nat Commun, 11, 2020
|
|
6LCM
 
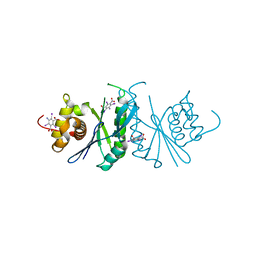 | | Crystal structure of chloroplast resolvase ZmMOC1 with the magic triangle I3C | | 分子名称: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, ZmMoc1 | | 著者 | Yan, J.J, Hong, S.X, Guan, Z.Y, Yin, P. | | 登録日 | 2019-11-19 | | 公開日 | 2020-04-08 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural insights into sequence-dependent Holliday junction resolution by the chloroplast resolvase MOC1.
Nat Commun, 11, 2020
|
|
6J15
 
 | | Complex structure of GY-5 Fab and PD-1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GY-5 heavy chain Fab, ... | | 著者 | Chen, D, Tan, S, Zhang, H, Wang, H, Chai, Y, Qi, J, Yan, J, Gao, G.F. | | 登録日 | 2018-12-27 | | 公開日 | 2019-11-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The FG Loop of PD-1 Serves as a "Hotspot" for Therapeutic Monoclonal Antibodies in Tumor Immune Checkpoint Therapy.
Iscience, 14, 2019
|
|
4XEM
 
 | |
4XEO
 
 | |
7C2Q
 
 | | The crystal structure of COVID-19 main protease in the apo state | | 分子名称: | 3C-like proteinase | | 著者 | Zhou, X.L, Zhong, F.L, Lin, C, Hu, X.H, Zhou, H, Wang, Q.S, Li, j, Zhang, J. | | 登録日 | 2020-05-08 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Structure of SARS-CoV-2 main protease in the apo state.
Sci China Life Sci, 64, 2021
|
|
