6E50
 
 | |
6E5E
 
 | |
6E5Q
 
 | |
6E7M
 
 | |
6E5R
 
 | |
3LST
 
 | | Crystal Structure of CalO1, Methyltransferase in Calicheamicin Biosynthesis, SAH bound form | | 分子名称: | 1,2-ETHANEDIOL, CalO1 Methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Chang, A, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | 登録日 | 2010-02-12 | | 公開日 | 2010-03-02 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural characterization of CalO1: a putative orsellinic acid methyltransferase in the calicheamicin-biosynthetic pathway.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
7MR8
 
 | |
6E6L
 
 | |
1NJM
 
 | | The crystal structure of the 50S Large ribosomal subunit from Deinococcus radiodurans complexed with a tRNA acceptor stem mimic (ASM) and the antibiotic sparsomycin | | 分子名称: | 23S ribosomal RNA, 50S ribosomal protein L16, GENERAL STRESS PROTEIN CTC, ... | | 著者 | Bashan, A, Agmon, I, Zarivatch, R, Schluenzen, F, Harms, J.M, Berisio, R, Bartels, H, Hansen, H.A, Yonath, A. | | 登録日 | 2003-01-02 | | 公開日 | 2003-02-11 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Structural basis of the ribosomal machinery for Peptide bond formation,
translocation, and nascent chain progression
Mol.Cell, 11, 2003
|
|
1NJO
 
 | | The crystal structure of the 50S Large ribosomal subunit from Deinococcus radiodurans complexed with a short substrate analog ACCPuromycin (ACCP) | | 分子名称: | 23S ribosomal RNA, RNA ACC(Puromycin) | | 著者 | Bashan, A, Agmon, I, Zarivatch, R, Schluenzen, F, Harms, J.M, Berisio, R, Bartels, H, Hansen, H.A, Yonath, A. | | 登録日 | 2003-01-02 | | 公開日 | 2003-02-11 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Structural basis of the ribosomal machinery for Peptide bond formation,
translocation, and nascent chain progression
Mol.Cell, 11, 2003
|
|
3OTG
 
 | | Crystal Structure of CalG1, Calicheamicin Glycostyltransferase, TDP bound form | | 分子名称: | CHLORIDE ION, CalG1, THYMIDINE-5'-DIPHOSPHATE | | 著者 | Chang, A, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2010-09-11 | | 公開日 | 2010-12-15 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3OTH
 
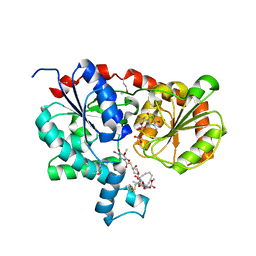 | | Crystal Structure of CalG1, Calicheamicin Glycostyltransferase, TDP and calicheamicin alpha3I bound form | | 分子名称: | CalG1, Calicheamicin alpha3I, THYMIDINE-5'-DIPHOSPHATE | | 著者 | Chang, A, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2010-09-11 | | 公開日 | 2010-12-15 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.301 Å) | | 主引用文献 | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3OTI
 
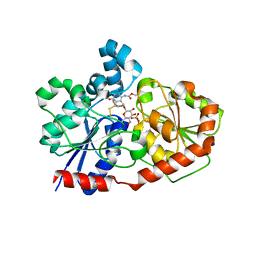 | | Crystal Structure of CalG3, Calicheamicin Glycostyltransferase, TDP and calicheamicin T0 bound form | | 分子名称: | CHLORIDE ION, CalG3, Calicheamicin T0, ... | | 著者 | Chang, A, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2010-09-11 | | 公開日 | 2010-12-15 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.597 Å) | | 主引用文献 | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6MOW
 
 | |
6NOE
 
 | |
6VID
 
 | |
6NNX
 
 | |
6NNY
 
 | |
7Z2J
 
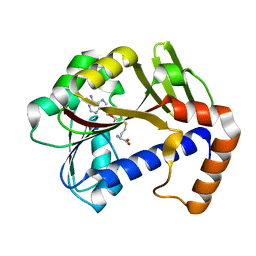 | | White Bream virus N7-Methyltransferase | | 分子名称: | Non-structural protein 1, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Shannon, A, Gauffre, P, Canard, B, Ferron, F. | | 登録日 | 2022-02-28 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.657 Å) | | 主引用文献 | A second type of N7-guanine RNA cap methyltransferase in an unusual locus of a large RNA virus genome.
Nucleic Acids Res., 50, 2022
|
|
1Q09
 
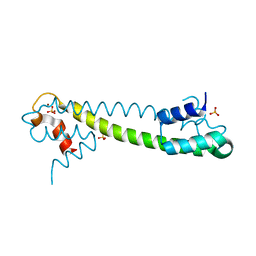 | | Crystal structure of the Zn(II) form of E. coli ZntR, a zinc-sensing transcriptional regulator (space group I4122) | | 分子名称: | SULFATE ION, ZINC ION, Zn(II)-responsive regulator of zntA | | 著者 | Changela, A, Chen, K, Xue, Y, Holschen, J, Outten, C.E, O'Halloran, T.V, Mondragon, A. | | 登録日 | 2003-07-15 | | 公開日 | 2003-09-16 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR
Science, 301, 2003
|
|
5CKW
 
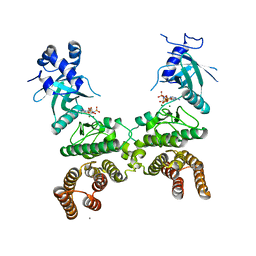 | | Crystal structure of LegK4_AMPPNP Kinase | | 分子名称: | CALCIUM ION, LegK4, MAGNESIUM ION, ... | | 著者 | Flayhan, A, Terradot, L. | | 登録日 | 2015-07-15 | | 公開日 | 2015-10-14 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | The structure of Legionella pneumophila LegK4 type four secretion system (T4SS) effector reveals a novel dimeric eukaryotic-like kinase.
Sci Rep, 5, 2015
|
|
1Q06
 
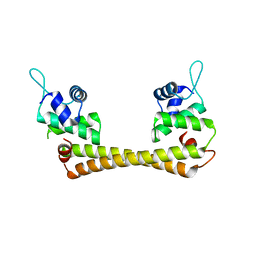 | | Crystal structure of the Ag(I) form of E. coli CueR, a copper efflux regulator | | 分子名称: | SILVER ION, Transcriptional regulator cueR | | 著者 | Changela, A, Chen, K, Xue, Y, Holschen, J, Outten, C.E, O'Halloran, T.V, Mondragon, A. | | 登録日 | 2003-07-15 | | 公開日 | 2003-09-16 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR
Science, 301, 2003
|
|
1Q0A
 
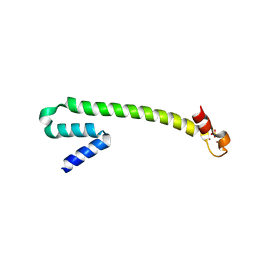 | | Crystal structure of the Zn(II) form of E. coli ZntR, a zinc-sensing transcriptional regulator (space group C222) | | 分子名称: | SULFATE ION, ZINC ION, Zn(II)-responsive regulator of zntA | | 著者 | Changela, A, Chen, K, Xue, Y, Holschen, J, Outten, C.E, O'Halloran, T.V, Mondragon, A. | | 登録日 | 2003-07-15 | | 公開日 | 2003-09-16 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR
Science, 301, 2003
|
|
1Q05
 
 | | Crystal structure of the Cu(I) form of E. coli CueR, a copper efflux regulator | | 分子名称: | COPPER (I) ION, Transcriptional regulator cueR | | 著者 | Changela, A, Chen, K, Xue, Y, Holschen, J, Outten, C.E, O'Halloran, T.V, Mondragon, A. | | 登録日 | 2003-07-15 | | 公開日 | 2003-09-16 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR
Science, 301, 2003
|
|
7Z05
 
 | | White Bream virus N7-Methyltransferase | | 分子名称: | DI(HYDROXYETHYL)ETHER, Non-structural protein 1, S-ADENOSYL-L-HOMOCYSTEINE, ... | | 著者 | Shannon, A, Gauffre, P, Canard, B, Ferron, F. | | 登録日 | 2022-02-22 | | 公開日 | 2022-09-28 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | A second type of N7-guanine RNA cap methyltransferase in an unusual locus of a large RNA virus genome.
Nucleic Acids Res., 50, 2022
|
|
