5ZDG
 
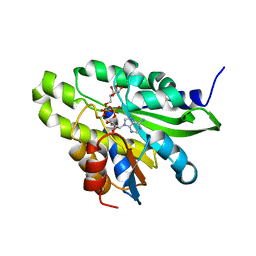 | | Crystal structure of poly(ADP-ribose) glycohydrolase (PARG) T267R mutant from Deinococcus radiodurans in complex with ADP-ribose | | 分子名称: | Poly APD-ribose glycohydrolase, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | 著者 | Cho, C.C, Hsu, C.H. | | 登録日 | 2018-02-23 | | 公開日 | 2019-02-27 | | 最終更新日 | 2020-03-11 | | 実験手法 | X-RAY DIFFRACTION (2.594 Å) | | 主引用文献 | Structural and biochemical evidence supporting poly ADP-ribosylation in the bacterium Deinococcus radiodurans.
Nat Commun, 10, 2019
|
|
5ZDE
 
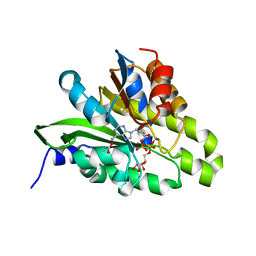 | | Crystal structure of poly(ADP-ribose) glycohydrolase (PARG) from Deinococcus radiodurans in complex with ADP-ribose (P3221) | | 分子名称: | Poly ADP-ribose glycohydrolase, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | 著者 | Cho, C.C, Hsu, C.H. | | 登録日 | 2018-02-23 | | 公開日 | 2019-02-27 | | 最終更新日 | 2020-03-11 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural and biochemical evidence supporting poly ADP-ribosylation in the bacterium Deinococcus radiodurans.
Nat Commun, 10, 2019
|
|
5ZDD
 
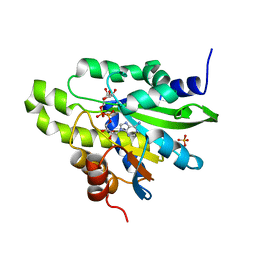 | | Crystal structure of poly(ADP-ribose) glycohydrolase (PARG) from Deinococcus radiodurans in complex with ADP-ribose (P212121) | | 分子名称: | PHOSPHATE ION, Poly ADP-ribose glycohydrolase, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | 著者 | Cho, C.C, Hsu, C.H. | | 登録日 | 2018-02-23 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.725 Å) | | 主引用文献 | Structural and biochemical evidence supporting poly ADP-ribosylation in the bacterium Deinococcus radiodurans.
Nat Commun, 10, 2019
|
|
5ZDF
 
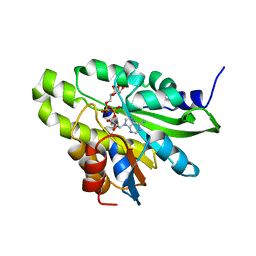 | | Crystal structure of poly(ADP-ribose) glycohydrolase (PARG) T267K mutant from Deinococcus radiodurans in complex with ADP-ribose | | 分子名称: | Poly ADP-ribose glycohydrolase, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | 著者 | Cho, C.C, Hsu, C.H. | | 登録日 | 2018-02-23 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.504 Å) | | 主引用文献 | Structural and biochemical evidence supporting poly ADP-ribosylation in the bacterium Deinococcus radiodurans.
Nat Commun, 10, 2019
|
|
7VE4
 
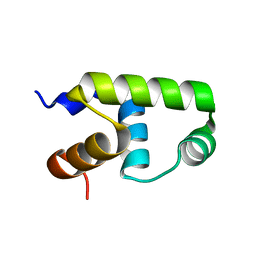 | | C-terminal domain of VraR | | 分子名称: | DNA-binding response regulator | | 著者 | Kumar, J.V, Chen, C, Hsu, C.H. | | 登録日 | 2021-09-08 | | 公開日 | 2022-05-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Structural insights into DNA binding domain of vancomycin-resistance-associated response regulator in complex with its promoter DNA from Staphylococcus aureus.
Protein Sci., 31, 2022
|
|
7VE6
 
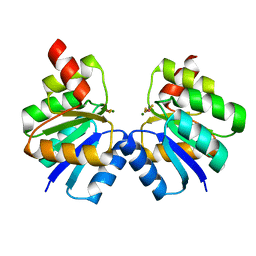 | | N-terminal domain of VraR | | 分子名称: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Response regulator protein VraR | | 著者 | Kumar, J.V, Chen, C, Hsu, C.H. | | 登録日 | 2021-09-08 | | 公開日 | 2022-05-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.77 Å) | | 主引用文献 | Structural insights into DNA binding domain of vancomycin-resistance-associated response regulator in complex with its promoter DNA from Staphylococcus aureus.
Protein Sci., 31, 2022
|
|
7VE5
 
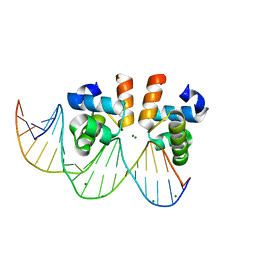 | | C-terminal domain of VraR | | 分子名称: | DNA-binding response regulator, MAGNESIUM ION, R1-DNA | | 著者 | Kumar, J.V, Chen, C, Hsu, C.H. | | 登録日 | 2021-09-08 | | 公開日 | 2022-05-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural insights into DNA binding domain of vancomycin-resistance-associated response regulator in complex with its promoter DNA from Staphylococcus aureus.
Protein Sci., 31, 2022
|
|
6JX7
 
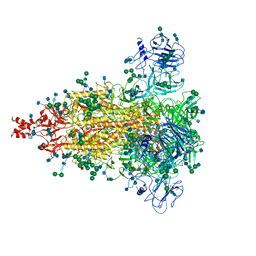 | | Cryo-EM structure of spike protein of feline infectious peritonitis virus strain UU4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Feline Infectious Peritonitis Virus Spike Protein, ... | | 著者 | Hsu, S.T.D, Yang, T.J, Ko, T.P, Draczkowski, P. | | 登録日 | 2019-04-22 | | 公開日 | 2020-01-15 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.31 Å) | | 主引用文献 | Cryo-EM analysis of a feline coronavirus spike protein reveals a unique structure and camouflaging glycans.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8HNK
 
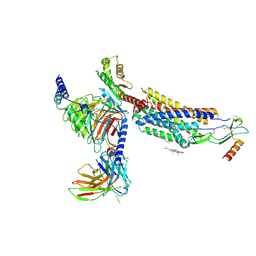 | | CXCR3-DNGi complex activated by CXCL11 | | 分子名称: | C-X-C motif chemokine 11, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Jiao, H.Z, Hu, H.L. | | 登録日 | 2022-12-08 | | 公開日 | 2023-11-29 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.01 Å) | | 主引用文献 | Structural insights into the activation and inhibition of CXC chemokine receptor 3.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HNN
 
 | | Structure of CXCR3 complexed with antagonist SCH546738 | | 分子名称: | 3-azanyl-6-chloranyl-5-[(3S)-4-[1-[(4-chlorophenyl)methyl]piperidin-4-yl]-3-ethyl-piperazin-1-yl]pyrazine-2-carboxamide, CHOLESTEROL, Nb6, ... | | 著者 | Jiao, H.Z, Hu, H.L. | | 登録日 | 2022-12-08 | | 公開日 | 2023-11-29 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural insights into the activation and inhibition of CXC chemokine receptor 3.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HNL
 
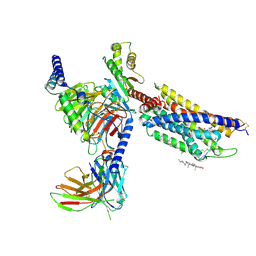 | | CXCR3-DNGi complex activated by PS372424 | | 分子名称: | (3S)-N-[(2S)-5-carbamimidamido-1-(cyclohexylmethylamino)-1-oxidanylidene-pentan-2-yl]-2-(4-oxidanylidene-4-phenyl-butanoyl)-3,4-dihydro-1H-isoquinoline-3-carboxamide, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Jiao, H.Z, Hu, H.L. | | 登録日 | 2022-12-08 | | 公開日 | 2023-11-29 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.98 Å) | | 主引用文献 | Structural insights into the activation and inhibition of CXC chemokine receptor 3.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HNM
 
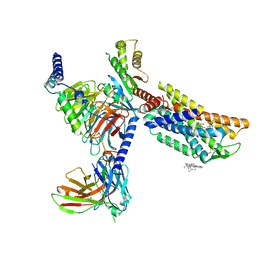 | | CXCR3-DNGi complex activated by VUF11222 | | 分子名称: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Jiao, H.Z, Hu, H.L. | | 登録日 | 2022-12-08 | | 公開日 | 2023-11-29 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.94 Å) | | 主引用文献 | Structural insights into the activation and inhibition of CXC chemokine receptor 3.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7C4H
 
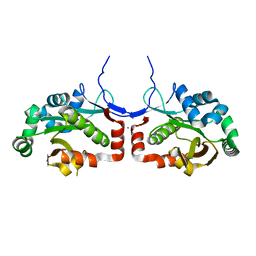 | |
7C33
 
 | |
7CZ4
 
 | |
6LXM
 
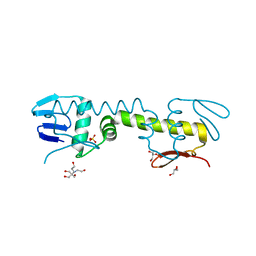 | | Crystal structure of C-terminal DNA-binding domain of Escherichia coli OmpR as a domain-swapped dimer | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, SULFATE ION, ... | | 著者 | Sadotra, S, Chen, C, Hsu, C.H. | | 登録日 | 2020-02-11 | | 公開日 | 2020-12-23 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.412 Å) | | 主引用文献 | Structural basis for promoter DNA recognition by the response regulator OmpR.
J.Struct.Biol., 213, 2020
|
|
6LXN
 
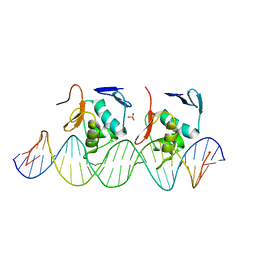 | |
6LXL
 
 | |
