5MVP
 
 | | Crystal structure of an A-DNA dodecamer containing the GGGCCC motif | | 分子名称: | DNA, POTASSIUM ION | | 著者 | Hardwick, J.S, Ptchelkine, D, Phillips, S.E.V, Brown, T. | | 登録日 | 2017-01-17 | | 公開日 | 2017-05-10 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.606 Å) | | 主引用文献 | 5-Formylcytosine does not change the global structure of DNA.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5MVK
 
 | |
5MVL
 
 | | Crystal structure of an A-DNA dodecamer containing 5-bromouracil | | 分子名称: | Brominated DNA dodecamer, MAGNESIUM ION | | 著者 | Hardwick, J.S, Ptchelkine, D, Phillips, S.E.V, Brown, T. | | 登録日 | 2017-01-16 | | 公開日 | 2017-05-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.405 Å) | | 主引用文献 | 5-Formylcytosine does not change the global structure of DNA.
Nat. Struct. Mol. Biol., 24, 2017
|
|
6QJR
 
 | | Crystal structure of a DNA dodecamer containing a tetramethylpyrrolinoxyl (nitroxide) spin label | | 分子名称: | DNA (5'-D(*CP*GP*CP*AP*AP*AP*AP*AP*AP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*TP*TP*TP*TP*TP*(5NO)P*GP*CP*G)-3') | | 著者 | Hardwick, J.S, Haugland, M.M, Ptchelkine, D, Brown, T, Anderson, E.E. | | 登録日 | 2019-01-24 | | 公開日 | 2020-02-12 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | 2'-Alkynyl spin-labelling is a minimally perturbing tool for DNA structural analysis.
Nucleic Acids Res., 48, 2020
|
|
4C64
 
 | | ULTRA HIGH RESOLUTION DICKERSON-DREW DODECAMER B-DNA | | 分子名称: | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*GP)-3', MAGNESIUM ION | | 著者 | Mcdonough, M, El-Sagheer, A.H, Brown, T, Schofield, C.J. | | 登録日 | 2013-09-17 | | 公開日 | 2013-10-02 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | Structural insights into how 5-hydroxymethylation influences transcription factor binding.
Chem. Commun. (Camb.), 50, 2014
|
|
4C5X
 
 | | Ultra High Resolution Dickerson-Drew dodecamer B-DNA with 5-Hydroxymethyl-cytosine Modification | | 分子名称: | 5'-D(CP*GP*CP*GP*AP*AP*TP*TP*5HCP*GP*CP*GP)-3', MAGNESIUM ION | | 著者 | McDonough, M, El-Sagheer, A.H, Brown, T, Schofield, C.J. | | 登録日 | 2013-09-16 | | 公開日 | 2013-10-02 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Structural insights into how 5-hydroxymethylation influences transcription factor binding.
Chem. Commun. (Camb.), 50, 2014
|
|
4C63
 
 | | ULTRA HIGH RESOLUTION DICKERSON-DREW DODECAMER B-DNA WITH 5- METHYLCYSTOSINE MODIFICATION | | 分子名称: | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*5CMP*GP*CP*GP)-3', MAGNESIUM ION | | 著者 | Mcdonough, M, El-Sagheer, A.H, Brown, T, Schofield, C.J. | | 登録日 | 2013-09-17 | | 公開日 | 2013-10-02 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | Structural insights into how 5-hydroxymethylation influences transcription factor binding.
Chem. Commun. (Camb.), 50, 2014
|
|
404D
 
 | |
2ANA
 
 | |
2X71
 
 | | Structural basis for the interaction of lactivicins with serine beta- lactamases | | 分子名称: | (2E)-2-{[(2S)-2-(ACETYLAMINO)-2-CARBOXYETHOXY]IMINO}PENTANEDIOIC ACID, BETA-LACTAMASE, ETHANOL, ... | | 著者 | Sauvage, E, Herman, R, Kerff, F, Rocaboy, M, Charlier, P. | | 登録日 | 2010-02-22 | | 公開日 | 2010-07-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Basis for the Interaction of Lactivicins with Serine Beta-Lactamases.
J.Med.Chem., 53, 2010
|
|
7NRO
 
 | | Crystal structure of AlkB in complex with manganese and N-(4-((6-((carboxymethyl)carbamoyl)-5-hydroxypyridin-2-yl)amino)phenyl)-N-oxohydroxylammonium | | 分子名称: | 2-[[6-[(4-nitrophenyl)amino]-3-oxidanyl-pyridin-2-yl]carbonylamino]ethanoic acid, Alpha-ketoglutarate-dependent dioxygenase AlkB, MANGANESE (II) ION | | 著者 | Shishodia, S, Maheswaran, P, Leissing, T, Aik, W.S, McDonough, M.A, Schofield, C.J. | | 登録日 | 2021-03-04 | | 公開日 | 2021-10-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Structure-Based Design of Selective Fat Mass and Obesity Associated Protein (FTO) Inhibitors.
J.Med.Chem., 64, 2021
|
|
1LAU
 
 | | URACIL-DNA GLYCOSYLASE | | 分子名称: | DNA (5'-D(*TP*TP*T)-3'), PROTEIN (URACIL-DNA GLYCOSYLASE (E.C.3.2.2.-)) | | 著者 | Pearl, L.H, Savva, R. | | 登録日 | 1996-01-03 | | 公開日 | 1996-06-10 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The structural basis of specific base-excision repair by uracil-DNA glycosylase.
Nature, 373, 1995
|
|
1D63
 
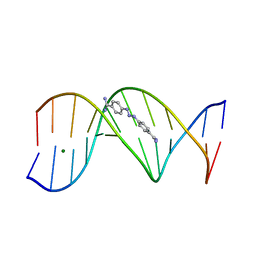 | | CRYSTAL STRUCTURE OF A BERENIL-D(CGCAAATTTGCG) COMPLEX; AN EXAMPLE OF DRUG-DNA RECOGNITION BASED ON SEQUENCE-DEPENDENT STRUCTURAL FEATURES | | 分子名称: | BERENIL, DNA (5'-D(*CP*GP*CP*AP*AP*AP*TP*TP*TP*GP*CP*G)-3'), MAGNESIUM ION | | 著者 | Brown, D.G, Sanderson, M.R, Garman, E, Neidle, S. | | 登録日 | 1992-03-02 | | 公開日 | 1993-01-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of a berenil-d(CGCAAATTTGCG) complex. An example of drug-DNA recognition based on sequence-dependent structural features.
J.Mol.Biol., 226, 1992
|
|
7E8Z
 
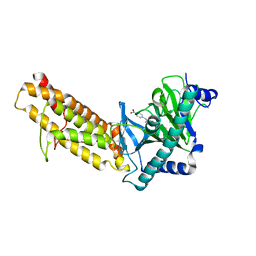 | | Crystal structure of the human fat mass and obesity associated protein (FTO) in complex with SS81 | | 分子名称: | 2-[[6-[(4-nitrophenyl)amino]-3-oxidanyl-pyridin-2-yl]carbonylamino]ethanoic acid, Alpha-ketoglutarate-dependent dioxygenase FTO, ZINC ION | | 著者 | Tam, N.Y, Ng, Y.M, Shishodia, S, McDonough, M.A, Schofield, C.J, Aik, W.S. | | 登録日 | 2021-03-03 | | 公開日 | 2021-10-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structure-Based Design of Selective Fat Mass and Obesity Associated Protein (FTO) Inhibitors.
J.Med.Chem., 64, 2021
|
|
1UDH
 
 | |
1UDG
 
 | |
4QHO
 
 | | Crystal structure of the human fat mass and obesity associated protein (FTO) in complex with CCO10 | | 分子名称: | Alpha-ketoglutarate-dependent dioxygenase FTO, N-{[3-hydroxy-6-(naphthalen-1-yl)pyridin-2-yl]carbonyl}glycine, ZINC ION | | 著者 | Aik, W.S, Clunie-O'Connor, C, McDonough, M.A, Schofield, C.J. | | 登録日 | 2014-05-28 | | 公開日 | 2015-06-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | Structure-Based Design of Selective Fat Mass and Obesity Associated Protein (FTO) Inhibitors.
J.Med.Chem., 64, 2021
|
|
2WPX
 
 | | Tandem GNAT protein from the clavulanic acid biosynthesis pathway (with AcCoA) | | 分子名称: | ACETYL COENZYME *A, GLYCEROL, ORF14 | | 著者 | Iqbal, A, Arunlanantham, H, McDonough, M.A, Chowdhury, R, Clifton, I.J. | | 登録日 | 2009-08-11 | | 公開日 | 2009-12-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Crystallographic and mass spectrometric analyses of a tandem GNAT protein from the clavulanic acid biosynthesis pathway.
Proteins, 78, 2010
|
|
2WPW
 
 | | Tandem GNAT protein from the clavulanic acid biosynthesis pathway (without AcCoA) | | 分子名称: | ACETYL COENZYME *A, ORF14 | | 著者 | Iqbal, A, Arunlanantham, H, McDonough, M.A, Chowdhury, R, Clifton, I.J. | | 登録日 | 2009-08-11 | | 公開日 | 2009-12-29 | | 最終更新日 | 2018-03-28 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Crystallographic and mass spectrometric analyses of a tandem GNAT protein from the clavulanic acid biosynthesis pathway.
Proteins, 78, 2010
|
|
1AT4
 
 | |
1AO9
 
 | |
1LVL
 
 | |
4II9
 
 | | Crystal structure of Weissella viridescens FemXVv non-ribosomal amino acid transferase in complex with a peptidyl-RNA conjugate | | 分子名称: | 5-mer peptide, FemX, GLYCEROL, ... | | 著者 | Li de la Sierra-Gallay, I, Fonvielle, M, van Tilbeurgh, H, Arthur, M, Etheve-Quelquejeu, M. | | 登録日 | 2012-12-20 | | 公開日 | 2013-07-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | The Structure of FemXWv in Complex with a Peptidyl-RNA Conjugate: Mechanism of Aminoacyl Transfer from Ala-tRNA(Ala) to Peptidoglycan Precursors
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
1UDI
 
 | |
