4L93
 
 | | Crystal structure of Human Hsp90 with S36 | | 分子名称: | 3,4-dihydroisoquinolin-2(1H)-yl[2,4-dihydroxy-5-(propan-2-yl)phenyl]methanone, Heat shock protein HSP 90-alpha | | 著者 | Li, J, Ren, J, Yang, M, Xiong, B, He, J. | | 登録日 | 2013-06-18 | | 公開日 | 2014-06-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.845 Å) | | 主引用文献 | Crystal structure of Human Hsp90 with S36
To be Published
|
|
4L90
 
 | | Crystal structure of Human Hsp90 with RL3 | | 分子名称: | Heat shock protein HSP 90-alpha, [5-(6-bromo[1,2,4]triazolo[4,3-a]pyridin-3-yl)-2,4-dihydroxyphenyl](4-methylpiperazin-1-yl)methanone | | 著者 | Li, J, Ren, J, Yang, M, Xiong, B, He, J. | | 登録日 | 2013-06-18 | | 公開日 | 2014-06-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.001 Å) | | 主引用文献 | Crystal structure of Human Hsp90 with RL3
to be published
|
|
2A7R
 
 | | Crystal structure of human Guanosine Monophosphate reductase 2 (GMPR2) | | 分子名称: | GMP reductase 2, GUANOSINE-5'-MONOPHOSPHATE, SULFATE ION | | 著者 | Li, J, Wei, Z, Zheng, M, Gu, X, Deng, Y, Qiu, R, Chen, F, Ji, C, Gong, W, Xie, Y, Mao, Y. | | 登録日 | 2005-07-05 | | 公開日 | 2006-01-31 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal Structure of Human Guanosine Monophosphate Reductase 2 (GMPR2) in Complex with GMP
J.Mol.Biol., 355, 2006
|
|
2AD7
 
 | | crystal structure of methanol dehydrogenase from M. W3A1 (form C) in the presence of methanol | | 分子名称: | CALCIUM ION, Methanol dehydrogenase subunit 1, Methanol dehydrogenase subunit 2, ... | | 著者 | Li, J, Gan, J.-H, Xia, Z.-X, Mathews, F.S. | | 登録日 | 2005-07-20 | | 公開日 | 2006-07-25 | | 最終更新日 | 2013-09-18 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The enzymatic reaction-induced configuration change of the prosthetic group PQQ of methanol dehydrogenase
Biochem.Biophys.Res.Commun., 406, 2011
|
|
2AD6
 
 | | crystal structure of methanol dehydrogenase from M. W3A1 (form C) | | 分子名称: | CALCIUM ION, Methanol dehydrogenase subunit 1, Methanol dehydrogenase subunit 2, ... | | 著者 | Li, J, Gan, J.-H, Xia, Z.-X, Mathews, F.S. | | 登録日 | 2005-07-20 | | 公開日 | 2006-07-25 | | 最終更新日 | 2013-09-18 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The enzymatic reaction-induced configuration change of the prosthetic group PQQ of methanol dehydrogenase
Biochem.Biophys.Res.Commun., 406, 2011
|
|
2AD8
 
 | | crystal structure of methanol dehydrogenase from M. W3A1 (form C) in the presence of ethanol | | 分子名称: | CALCIUM ION, Methanol dehydrogenase subunit 1, Methanol dehydrogenase subunit 2, ... | | 著者 | Li, J, Gan, J.-H, Xia, Z.-X, Mathews, F.S. | | 登録日 | 2005-07-20 | | 公開日 | 2006-07-25 | | 最終更新日 | 2013-09-18 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The enzymatic reaction-induced configuration change of the prosthetic group PQQ of methanol dehydrogenase
Biochem.Biophys.Res.Commun., 406, 2011
|
|
6L8W
 
 | | Crystal structure of ugt transferase mutant2 | | 分子名称: | Glycosyltransferase | | 著者 | Li, J, Shan, N, Yang, J.G, Liu, W.D, Sun, Y.X. | | 登録日 | 2019-11-07 | | 公開日 | 2020-04-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Efficient O-Glycosylation of Triterpenes Enabled by Protein Engineering of Plant Glycosyltransferase UGT74AC1
Acs Catalysis, 2020
|
|
6L90
 
 | | Crystal structure of ugt transferase enzyme | | 分子名称: | Glycosyltransferase, SULFATE ION | | 著者 | Li, J, Shan, N, Yang, J.G, Liu, W.D, Sun, Y.X. | | 登録日 | 2019-11-07 | | 公開日 | 2020-04-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Efficient O-Glycosylation of Triterpenes Enabled by Protein Engineering of Plant Glycosyltransferase UGT74AC1
Acs Catalysis, 2020
|
|
6L8X
 
 | | Crystal structure of Siraitia grosvenorii ugt transferase mutant2 | | 分子名称: | Glycosyltransferase | | 著者 | Li, J, Shan, N, Yang, J.G, Liu, W.D, Sun, Y.X. | | 登録日 | 2019-11-07 | | 公開日 | 2020-04-08 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Efficient O-Glycosylation of Triterpenes Enabled by Protein Engineering of Plant Glycosyltransferase UGT74AC1
Acs Catalysis, 10, 2020
|
|
5CF0
 
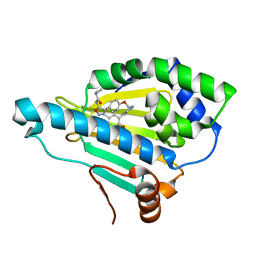 | | Crystal Structure of the human Hsp90-alpha N-domain bound to the hsp90 inhibitor FJ6 | | 分子名称: | Heat shock protein HSP 90-alpha, N-{3-[2,4-dihydroxy-5-(isoquinolin-4-yl)phenyl]-4-(4-methoxyphenyl)-1,2-oxazol-5-yl}cyclopropanecarboxamide | | 著者 | Li, J, Shi, F, Xiong, B, He, J.H. | | 登録日 | 2015-07-08 | | 公開日 | 2016-07-13 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | FS23 binds to the N-terminal domain of human Hsp90: A novel small inhibitor for Hsp90
Nucl.Sci.Tech., 26, 2015
|
|
8X51
 
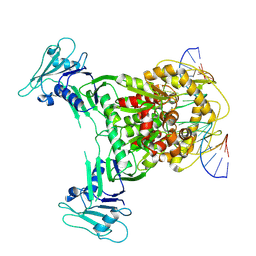 | | Cryo-EM structure of Gabija GajA in complex with DNA(focused refinement) | | 分子名称: | CALCIUM ION, DNA (5'-D(*AP*AP*AP*AP*AP*TP*AP*AP*CP*CP*GP*GP*GP*TP*TP*AP*TP*TP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*AP*TP*AP*AP*CP*CP*CP*GP*GP*TP*TP*AP*TP*TP*TP*TP*T)-3'), ... | | 著者 | Li, J, Wang, Z, Wang, L. | | 登録日 | 2023-11-16 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (2.92 Å) | | 主引用文献 | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
8WY5
 
 | | Structure of Gabija GajA in complex with DNA | | 分子名称: | CALCIUM ION, DNA (5'-D(P*AP*AP*AP*AP*TP*AP*AP*CP*CP*GP*GP*GP*TP*TP*AP*TP*TP*AP*A)-3'), DNA (5'-D(P*TP*TP*AP*AP*TP*AP*AP*CP*CP*CP*GP*GP*TP*TP*AP*TP*TP*TP*T)-3'), ... | | 著者 | Li, J, Wang, Z, Wang, L. | | 登録日 | 2023-10-30 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.12 Å) | | 主引用文献 | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
1SEK
 
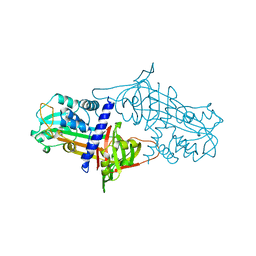 | | THE STRUCTURE OF ACTIVE SERPIN K FROM MANDUCA SEXTA AND A MODEL FOR SERPIN-PROTEASE COMPLEX FORMATION | | 分子名称: | SERPIN K | | 著者 | Li, J, Wang, Z, Canagarajah, B, Jiang, H, Kanost, M, Goldsmith, E.J. | | 登録日 | 1998-03-06 | | 公開日 | 1999-03-23 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The structure of active serpin 1K from Manduca sexta.
Structure Fold.Des., 7, 1999
|
|
8WY4
 
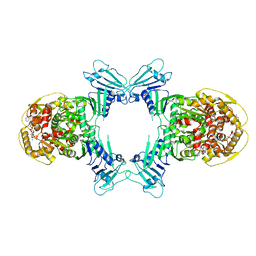 | | GajA tetramer with ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Endonuclease GajA | | 著者 | Li, J, Wang, Z, Wang, L. | | 登録日 | 2023-10-30 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (2.81 Å) | | 主引用文献 | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
8X5N
 
 | | Tetramer Gabija with ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Endonuclease GajA, Gabija protein GajB, ... | | 著者 | Li, J, Wang, Z, Wang, L. | | 登録日 | 2023-11-17 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structures and activation mechanism of the Gabija anti-phage system.
Nature, 629, 2024
|
|
8X5I
 
 | |
6JLE
 
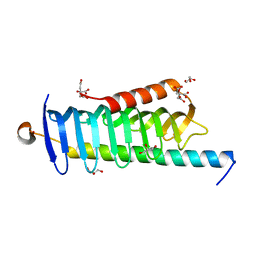 | | Crystal structure of MORN4/Myo3a complex | | 分子名称: | CITRIC ACID, GLYCEROL, MORN repeat-containing protein 4, ... | | 著者 | Li, J, Liu, H, Raval, M.H, Wan, J, Yengo, C.M, Liu, W, Zhang, M. | | 登録日 | 2019-03-05 | | 公開日 | 2019-07-24 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structure of the MORN4/Myo3a Tail Complex Reveals MORN Repeats as Protein Binding Modules.
Structure, 27, 2019
|
|
4JN9
 
 | | Crystal structure of the DepH | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, DepH, ... | | 著者 | Li, J, Wang, C, Zhang, Z.M, Zhou, J.H, Cheng, E. | | 登録日 | 2013-03-14 | | 公開日 | 2014-04-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The structural basis of an NADP+-independent dithiol oxidase in FK228 biosynthesis.
Sci Rep, 4, 2014
|
|
5ZVD
 
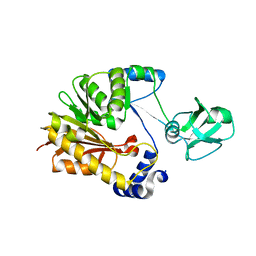 | |
5ZVG
 
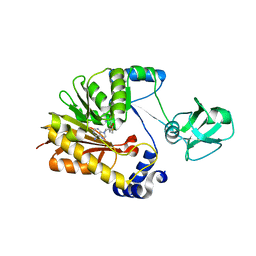 | |
5ZVE
 
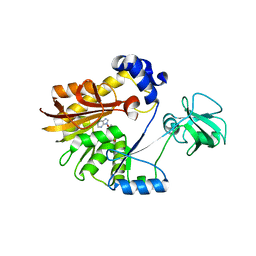 | |
5ZVH
 
 | |
3RY7
 
 | | Crystal Structure of Sa239 | | 分子名称: | GLYCEROL, Ribokinase | | 著者 | Li, J, Wu, M, Wang, L, Zang, J. | | 登録日 | 2011-05-11 | | 公開日 | 2012-04-25 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystal structure of Sa239 reveals the structural basis for the activation of ribokinase by monovalent cations.
J.Struct.Biol., 177, 2012
|
|
7WJS
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor Y13157 | | 分子名称: | 2-(2-cyclobutyl-1~{H}-imidazol-5-yl)-7-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-5-methyl-furo[3,2-c]pyridin-4-one, Bromodomain-containing protein 4, GLYCEROL | | 著者 | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | 登録日 | 2022-01-07 | | 公開日 | 2022-08-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.73 Å) | | 主引用文献 | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7WKY
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor Y13153 | | 分子名称: | 2-(2-cyclopentyl-1~{H}-imidazol-5-yl)-7-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-5-methyl-furo[3,2-c]pyridin-4-one, Bromodomain-containing protein 4, GLYCEROL | | 著者 | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | 登録日 | 2022-01-12 | | 公開日 | 2022-08-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.83 Å) | | 主引用文献 | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
