2AAT
 
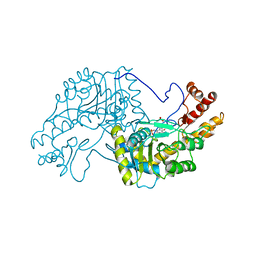 | | 2.8-ANGSTROMS-RESOLUTION CRYSTAL STRUCTURE OF AN ACTIVE-SITE MUTANT OF ASPARTATE AMINOTRANSFERASE FROM ESCHERICHIA COLI | | 分子名称: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ASPARTATE AMINOTRANSFERASE, SULFATE ION | | 著者 | Smith, D, Almo, S.C, Toney, M, Ringe, D. | | 登録日 | 1989-05-30 | | 公開日 | 1989-10-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | 2.8-A-resolution crystal structure of an active-site mutant of aspartate aminotransferase from Escherichia coli.
Biochemistry, 28, 1989
|
|
9GPB
 
 | |
1GWK
 
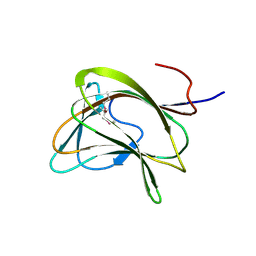 | | Carbohydrate binding module family29 | | 分子名称: | NON-CATALYTIC PROTEIN 1 | | 著者 | Charnock, S.J, Nurizzo, D, Davies, G.J. | | 登録日 | 2002-03-19 | | 公開日 | 2003-03-20 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Promiscuity in Ligand-Binding: The Three-Dimensional Structure of a Piromyces Carbohydrate-Binding Module,Cbm29-2,in Complex with Cello- and Mannohexaose
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1HCQ
 
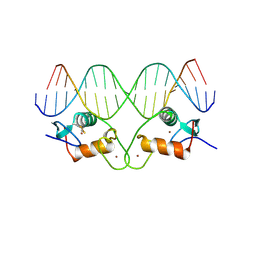 | | THE CRYSTAL STRUCTURE OF THE ESTROGEN RECEPTOR DNA-BINDING DOMAIN BOUND TO DNA: HOW RECEPTORS DISCRIMINATE BETWEEN THEIR RESPONSE ELEMENTS | | 分子名称: | DNA (5'-D(*CP*CP*AP*GP*GP*TP*CP*AP*CP*AP*GP*TP*GP*AP*CP*CP*T P*G)-3'), DNA (5'-D(*CP*CP*AP*GP*GP*TP*CP*AP*CP*TP*GP*TP*GP*AP*CP*CP*T P*G)-3'), PROTEIN (ESTROGEN RECEPTOR), ... | | 著者 | Schwabe, J.W.R, Chapman, L, Finch, J.T, Rhodes, D. | | 登録日 | 1995-01-04 | | 公開日 | 1995-11-23 | | 最終更新日 | 2022-12-21 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The crystal structure of the estrogen receptor DNA-binding domain bound to DNA: how receptors discriminate between their response elements.
Cell(Cambridge,Mass.), 75, 1993
|
|
1HCF
 
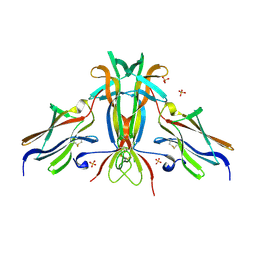 | | Crystal structure of TrkB-d5 bound to neurotrophin-4/5 | | 分子名称: | BDNF/NT-3 GROWTH FACTORS RECEPTOR, NEUROTROPHIN-4, SULFATE ION | | 著者 | Banfield, M.J, Naylor, R.L, Robertson, A.G.S, Allen, S.J, Dawbarn, D, Brady, R.L. | | 登録日 | 2001-05-03 | | 公開日 | 2001-12-06 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Specificity in Trk-Receptor:Neurotrophin Interaction: The Crystal Structure of Trkb-D5 in Complex with Neurotrophin-4/5
Structure, 9, 2001
|
|
1GWL
 
 | | Carbohydrate binding module family29 complexed with mannohexaose | | 分子名称: | NON-CATALYTIC PROTEIN 1, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | 著者 | Charnock, S.J, Nurizzo, D, Davies, G.J. | | 登録日 | 2002-03-19 | | 公開日 | 2003-03-20 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Promiscuity in Ligand-Binding: The Three-Dimensional Structure of a Piromyces Carbohydrate-Binding Module,Cbm29-2,in Complex with Cello- and Mannohexaose
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1GWM
 
 | | Carbohydrate binding module family29 complexed with glucohexaose | | 分子名称: | 1,2-ETHANEDIOL, COBALT (II) ION, NON-CATALYTIC PROTEIN 1, ... | | 著者 | Charnock, S.J, Nurizzo, D, Davies, G.J. | | 登録日 | 2002-03-19 | | 公開日 | 2003-03-20 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Promiscuity in Ligand-Binding: The Three-Dimensional Structure of a Piromyces Carbohydrate-Binding Module,Cbm29-2,in Complex with Cello- and Mannohexaose
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1GSG
 
 | | Structure of E.coli glutaminyl-tRNA synthetase complexed with trnagln and ATP at 2.8 Angstroms resolution | | 分子名称: | GLUTAMINYL-TRNA SYNTHETASE, TRNAGLN | | 著者 | Rould, M.A, Perona, J.J, Soell, D, Steitz, T.A. | | 登録日 | 1990-04-03 | | 公開日 | 1992-02-24 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP at 2.8 A resolution.
Science, 246, 1989
|
|
1PPH
 
 | | GEOMETRY OF BINDING OF THE NALPHA-TOSYLATED PIPERIDIDES OF M-AMIDINO-, P-AMIDINO-AND P-GUANIDINO PHENYLALANINE TO THROMBIN AND TRYPSIN: X-RAY CRYSTAL STRUCTURES OF THEIR TRYPSIN COMPLEXES AND MODELING OF THEIR THROMBIN COMPLEXES | | 分子名称: | 3-[(2S)-2-{[(4-methylphenyl)sulfonyl]amino}-3-oxo-3-piperidin-1-ylpropyl]benzenecarboximidamide, CALCIUM ION, SULFATE ION, ... | | 著者 | Bode, W, Turk, D. | | 登録日 | 1991-10-24 | | 公開日 | 1994-01-31 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Geometry of binding of the N alpha-tosylated piperidides of m-amidino-, p-amidino- and p-guanidino phenylalanine to thrombin and trypsin. X-ray crystal structures of their trypsin complexes and modeling of their thrombin complexes.
FEBS Lett., 287, 1991
|
|
6PLG
 
 | | Crystal structure of human PHGDH complexed with Compound 15 | | 分子名称: | (2S)-(4-{3-[(4,5-dichloro-1-methyl-1H-indole-2-carbonyl)amino]oxetan-3-yl}phenyl)(pyridin-3-yl)acetic acid, D-3-phosphoglycerate dehydrogenase, D-MALATE | | 著者 | Olland, A, Lakshminarasimhan, D, White, A, Suto, R.K. | | 登録日 | 2019-06-30 | | 公開日 | 2019-07-24 | | 最終更新日 | 2019-09-04 | | 実験手法 | X-RAY DIFFRACTION (2.93 Å) | | 主引用文献 | Inhibition of 3-phosphoglycerate dehydrogenase (PHGDH) by indole amides abrogates de novo serine synthesis in cancer cells.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
1QAJ
 
 | | CRYSTAL STRUCTURES OF THE N-TERMINAL FRAGMENT FROM MOLONEY MURINE LEUKEMIA VIRUS REVERSE TRANSCRIPTASE COMPLEXED WITH NUCLEIC ACID: FUNCTIONAL IMPLICATIONS FOR TEMPLATE-PRIMER BINDING TO THE FINGERS DOMAIN | | 分子名称: | DNA (5'-D(*CP*AP*TP*GP*CP*AP*TP*G)-3'), REVERSE TRANSCRIPTASE | | 著者 | Najmudin, S, Cote, M, Sun, D, Yohannan, S, Montano, S.P, Gu, J, Georgiadis, M.M. | | 登録日 | 1999-03-18 | | 公開日 | 2000-04-02 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structures of an N-terminal fragment from Moloney murine leukemia virus reverse transcriptase complexed with nucleic acid: functional implications for template-primer binding to the fingers domain.
J.Mol.Biol., 296, 2000
|
|
8C66
 
 | |
6P0J
 
 | | Crystal structure of GDP-bound human RalA | | 分子名称: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, Ras-related protein Ral-A | | 著者 | Bum-Erdene, K, Gonzalez-Gutierrez, G, Liu, D, Meroueh, S.O. | | 登録日 | 2019-05-17 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.31 Å) | | 主引用文献 | Small-molecule covalent bond formation at tyrosine creates a binding site and inhibits activation of Ral GTPases.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7SL5
 
 | |
7SKZ
 
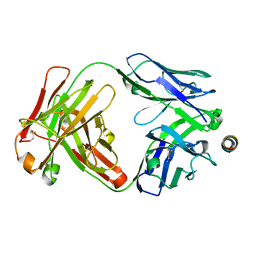 | |
6PBW
 
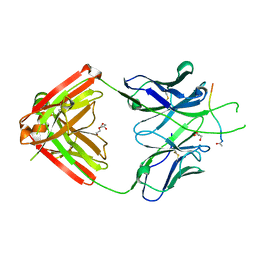 | | Crystal structure of Fab667 complex | | 分子名称: | Fab667 heavy chain, Fab667 light chain, GLYCEROL, ... | | 著者 | Oyen, D, Wilson, I.A. | | 登録日 | 2019-06-14 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.058 Å) | | 主引用文献 | Structure and mechanism of monoclonal antibody binding to the junctional epitope of Plasmodium falciparum circumsporozoite protein.
Plos Pathog., 16, 2020
|
|
7AK4
 
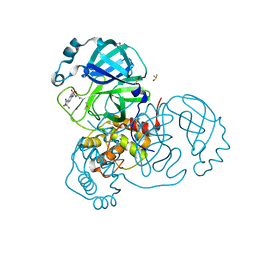 | | Structure of SARS-CoV-2 Main Protease bound to Tretazicar. | | 分子名称: | 3C-like proteinase, 5-(AZIRIDIN-1-YL)-2,4-DINITROBENZAMIDE, CHLORIDE ION, ... | | 著者 | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | 登録日 | 2020-09-29 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AMJ
 
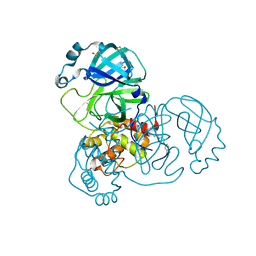 | | Structure of SARS-CoV-2 Main Protease bound to PD 168568. | | 分子名称: | (3~{S})-3-[2-[4-(3,4-dimethylphenyl)piperazin-1-yl]ethyl]-2,3-dihydroisoindol-1-one, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Hinrichs, W, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Boger, J, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | 登録日 | 2020-10-09 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AKU
 
 | | Structure of SARS-CoV-2 Main Protease bound to Calpeptin. | | 分子名称: | 3C-like proteinase, CHLORIDE ION, Calpeptin | | 著者 | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | 登録日 | 2020-10-02 | | 公開日 | 2020-10-28 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AX6
 
 | | Structure of SARS-CoV-2 Main Protease bound to Glutathione isopropyl ester | | 分子名称: | (2~{S})-2-azanyl-5-oxidanylidene-5-[[(2~{S})-1-oxidanylidene-1-[(2-oxidanylidene-2-propan-2-yloxy-ethyl)amino]-3-sulfanyl-propan-2-yl]amino]pentanoic acid, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | 著者 | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | 登録日 | 2020-11-09 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AWR
 
 | | Structure of SARS-CoV-2 Main Protease bound to Tegafur | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, TEGAFUR | | 著者 | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | 登録日 | 2020-11-09 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.34 Å) | | 主引用文献 | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7ARF
 
 | | Structure of SARS-CoV-2 Main Protease bound to thioglucose. | | 分子名称: | (2~{S},3~{R},4~{R},5~{S},6~{S})-2-(hydroxymethyl)-6-sulfanyl-oxane-3,4,5-triol, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | 登録日 | 2020-10-24 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AY7
 
 | | Structure of SARS-CoV-2 Main Protease bound to Isofloxythepin | | 分子名称: | 3C-like proteinase, 9-fluoranyl-3-propan-2-yl-5,6-dihydrobenzo[b][1]benzothiepine, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | 登録日 | 2020-11-11 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AR6
 
 | | Structure of apo SARS-CoV-2 Main Protease with large beta angle, space group C2. | | 分子名称: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE | | 著者 | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Hinrichs, W, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Boger, J, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | 登録日 | 2020-10-23 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AP6
 
 | | Structure of SARS-CoV-2 Main Protease bound to MUT056399. | | 分子名称: | 3C-like proteinase, 4-(4-ethyl-5-fluoranyl-2-oxidanyl-phenoxy)-3-fluoranyl-benzamide | | 著者 | Ewert, W, Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | 登録日 | 2020-10-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
