6ZJT
 
 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant E441Q in complex with lactulose | | 分子名称: | ACETATE ION, Beta-galactosidase, SODIUM ION, ... | | 著者 | Rutkiewicz, M, Bujacz, A, Bujacz, G. | | 登録日 | 2020-06-29 | | 公開日 | 2020-08-05 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Mapping the Transglycosylation Relevant Sites of Cold-Adapted beta-d-Galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 21, 2020
|
|
6ZJV
 
 | |
6ZJR
 
 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant E517Q in complex with lactulose | | 分子名称: | ACETATE ION, Beta-galactosidase, MALONATE ION, ... | | 著者 | Rutkiewicz, M, Bujacz, A, Bujacz, G. | | 登録日 | 2020-06-29 | | 公開日 | 2020-08-05 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Mapping the Transglycosylation Relevant Sites of Cold-Adapted beta-d-Galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 21, 2020
|
|
6ZJP
 
 | |
6ZJS
 
 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant E441Q in complex with galactose | | 分子名称: | (2S)-2-hydroxybutanedioic acid, ACETATE ION, Beta-galactosidase, ... | | 著者 | Rutkiewicz, M, Bujacz, A, Bujacz, G. | | 登録日 | 2020-06-29 | | 公開日 | 2020-08-05 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Mapping the Transglycosylation Relevant Sites of Cold-Adapted beta-d-Galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 21, 2020
|
|
4TTH
 
 | | Crystal structure of a CDK6/Vcyclin complex with inhibitor bound | | 分子名称: | 9-cyclopentyl-N-(5-piperazin-1-ylpyridin-2-yl)pyrido[4,5]pyrrolo[1,2-d]pyrimidin-2-amine, Cyclin homolog, Cyclin-dependent kinase 6 | | 著者 | Piper, D.E, Walker, N, Wang, Z. | | 登録日 | 2014-06-20 | | 公開日 | 2014-08-06 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Discovery of AMG 925, a FLT3 and CDK4 dual kinase inhibitor with preferential affinity for the activated state of FLT3.
J.Med.Chem., 57, 2014
|
|
9JE0
 
 | | Human URAT1 bound to benzbromarone | | 分子名称: | Solute carrier family 22 member 12, [3,5-bis(bromanyl)-4-oxidanyl-phenyl]-(2-ethyl-1-benzofuran-3-yl)methanone | | 著者 | Wu, C, Zhang, C, Jin, S, Wang, J.J, Dai, A, Jiang, Y, Yang, D, Xu, H.E. | | 登録日 | 2024-09-01 | | 公開日 | 2024-10-16 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.23 Å) | | 主引用文献 | Molecular mechanisms of uric acid transport by the native human URAT1 and its inhibition by anti-gout drugs
Biorxiv, 2024
|
|
9JE1
 
 | | Human URAT1 bound to dotinurad | | 分子名称: | Solute carrier family 22 member 12, dotinurad | | 著者 | Wu, C, Zhang, C, Jin, S, Wang, J.J, Dai, A, Jiang, Y, Yang, D, Xu, H.E. | | 登録日 | 2024-09-01 | | 公開日 | 2024-10-16 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Molecular mechanisms of uric acid transport by the native human URAT1 and its inhibition by anti-gout drugs
Biorxiv, 2024
|
|
9JDY
 
 | | Human URAT1 bound with verinurad | | 分子名称: | Solute carrier family 22 member 12, verinurad | | 著者 | Wu, C, Zhang, C, Jin, S, Wang, J.J, Dai, A, Jiang, Y, Yang, D, Xu, H.E. | | 登録日 | 2024-09-01 | | 公開日 | 2024-10-16 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.23 Å) | | 主引用文献 | Molecular mechanisms of uric acid transport by the native human URAT1 and its inhibition by anti-gout drugs
Biorxiv, 2024
|
|
9JDV
 
 | | Human URAT1 bound with Uric acid | | 分子名称: | Solute carrier family 22 member 12, URIC ACID | | 著者 | Wu, C, Zhang, C, Jin, S, Wang, J.J, Jiang, Y, Yang, D, Xu, H.E. | | 登録日 | 2024-09-01 | | 公開日 | 2024-10-16 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (3.32 Å) | | 主引用文献 | Molecular mechanisms of uric acid transport by the native human URAT1 and its inhibition by anti-gout drugs
Biorxiv, 2024
|
|
6N05
 
 | | Structure of anti-crispr protein, AcrIIC2 | | 分子名称: | AcrIIC2 | | 著者 | Shah, M, Thavalingham, A, Maxwell, K.L, Moraes, T.F. | | 登録日 | 2018-11-06 | | 公開日 | 2019-06-05 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Inhibition of CRISPR-Cas9 ribonucleoprotein complex assembly by anti-CRISPR AcrIIC2.
Nat Commun, 10, 2019
|
|
9JDZ
 
 | | Human URAT1 bound to lesinurad | | 分子名称: | Solute carrier family 22 member 12, lesinurad | | 著者 | Wu, C, Xu, H.E. | | 登録日 | 2024-09-01 | | 公開日 | 2024-10-16 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Molecular mechanisms of uric acid transport by the native human URAT1 and its inhibition by anti-gout drugs
Biorxiv, 2024
|
|
4L02
 
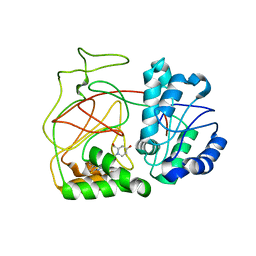 | | Crystal Structure of SphK1 with inhibitor | | 分子名称: | (2R,4S)-1-[2-(4-{[4-(3,4-dichlorophenyl)-1,3-thiazol-2-yl]amino}phenyl)ethyl]-2-(hydroxymethyl)piperidin-4-ol, Sphingosine kinase 1 | | 著者 | Min, X, Walker, N, Wang, Z. | | 登録日 | 2013-05-30 | | 公開日 | 2013-07-24 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structure guided design of a series of sphingosine kinase (SphK) inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
8YE4
 
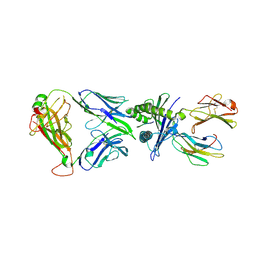 | | The complex of TCR NYN-I and HLA-A24 bound to SARS-CoV-2 Spike448-456 peptide NYNYLYRLF | | 分子名称: | Beta-2-microglobulin, MHC class I antigen precusor, Spike protein S1, ... | | 著者 | Deng, S.S, Jin, T.C, Xu, Z.H, Wang, M.H. | | 登録日 | 2024-02-21 | | 公開日 | 2024-08-21 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural insights into immune escape at killer T cell epitope by SARS-CoV-2 Spike Y453F variants.
J.Biol.Chem., 300, 2024
|
|
8ZV9
 
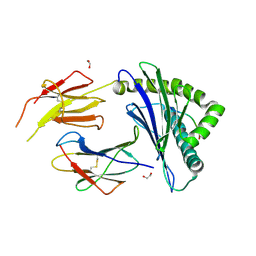 | | Complex structure of HLA2402 with recognizing SARS-CoV-2 Y453F epitope NYNYLFRLF | | 分子名称: | 1,2-ETHANEDIOL, Beta-2-microglobulin, MHC class I antigen, ... | | 著者 | Deng, S.S, Jin, T.C, Xu, Z.H, Wang, M.H. | | 登録日 | 2024-06-11 | | 公開日 | 2024-08-21 | | 最終更新日 | 2024-08-28 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural insights into immune escape at killer T cell epitope by SARS-CoV-2 Spike Y453F variants.
J.Biol.Chem., 300, 2024
|
|
3H0A
 
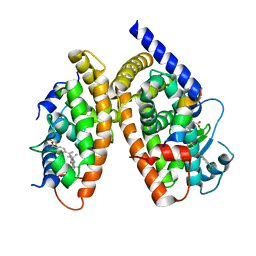 | | Crystal Structure of Peroxisome Proliferator-Activated Receptor Gamma (PPARg) and Retinoic Acid Receptor Alpha (RXRa) in Complex with 9-cis Retinoic Acid, Co-activator Peptide, and a Partial Agonist | | 分子名称: | 4-[1-(3,5,5,8,8-pentamethyl-5,6,7,8-tetrahydronaphthalen-2-yl)ethenyl]benzoic acid, Nuclear receptor coactivator 1, Co-activator Peptide, ... | | 著者 | Wang, Z, Sudom, A, Walker, N.P. | | 登録日 | 2009-04-08 | | 公開日 | 2009-06-09 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Identification of a PPARdelta agonist with partial agonistic activity on PPARgamma.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3GZ9
 
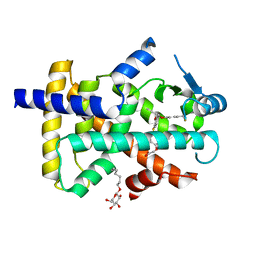 | | Crystal Structure of Peroxisome Proliferator-Activated Receptor Delta (PPARd) in Complex with a Full Agonist | | 分子名称: | (2,3-dimethyl-4-{[2-(prop-2-yn-1-yloxy)-4-{[4-(trifluoromethyl)phenoxy]methyl}phenyl]sulfanyl}phenoxy)acetic acid, Peroxisome proliferator-activated receptor delta, heptyl beta-D-glucopyranoside | | 著者 | Wang, Z, Sudom, A, Walker, N.P. | | 登録日 | 2009-04-06 | | 公開日 | 2009-06-30 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Identification of a PPARdelta agonist with partial agonistic activity on PPARgamma.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2OSH
 
 | |
8G83
 
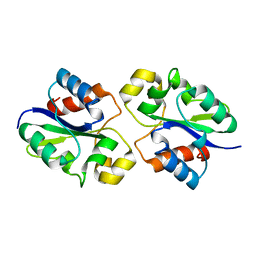 | | Structure of NAD+ consuming protein Acinetobacter baumannii TIR domain | | 分子名称: | NAD(+) hydrolase AbTIR | | 著者 | Klontz, E.H, Wang, Y, Glendening, G, Carr, J, Tsibouris, T, Buddula, S, Nallar, S, Soares, A, Snyder, G.A. | | 登録日 | 2023-02-17 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.03 Å) | | 主引用文献 | The structure of NAD + consuming protein Acinetobacter baumannii TIR domain shows unique kinetics and conformations.
J.Biol.Chem., 299, 2023
|
|
4LZO
 
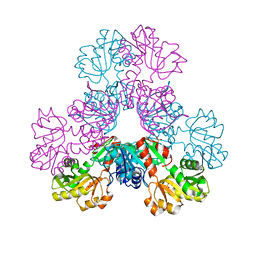 | | Crystal structure of human PRS1 A87T mutant | | 分子名称: | Ribose-phosphate pyrophosphokinase 1, SULFATE ION | | 著者 | Chen, P, Teng, M, Li, X. | | 登録日 | 2013-07-31 | | 公開日 | 2015-02-04 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.31 Å) | | 主引用文献 | Crystal and EM Structures of Human Phosphoribosyl Pyrophosphate Synthase I (PRS1) Provide Novel Insights into the Disease-Associated Mutations
Plos One, 10, 2015
|
|
4LYG
 
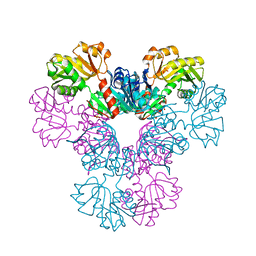 | | Crystal structure of human PRS1 E43T mutant | | 分子名称: | Ribose-phosphate pyrophosphokinase 1, SULFATE ION | | 著者 | Chen, P, Teng, M, Li, X. | | 登録日 | 2013-07-31 | | 公開日 | 2015-02-04 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal and EM Structures of Human Phosphoribosyl Pyrophosphate Synthase I (PRS1) Provide Novel Insights into the Disease-Associated Mutations
Plos One, 10, 2015
|
|
4LZN
 
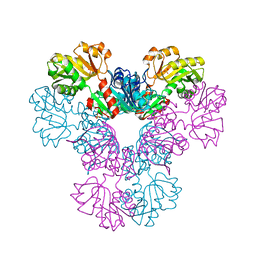 | | Crystal structure of human PRS1 D65N mutant | | 分子名称: | Ribose-phosphate pyrophosphokinase 1, SULFATE ION | | 著者 | Chen, P, Teng, M, Li, X. | | 登録日 | 2013-07-31 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Crystal and EM Structures of Human Phosphoribosyl Pyrophosphate Synthase I (PRS1) Provide Novel Insights into the Disease-Associated Mutations
Plos One, 10, 2015
|
|
4M0P
 
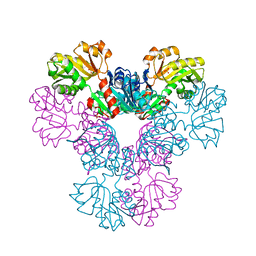 | | Crystal structure of human PRS1 M115T mutant | | 分子名称: | Ribose-phosphate pyrophosphokinase 1, SULFATE ION | | 著者 | Chen, P, Teng, M, Li, X. | | 登録日 | 2013-08-01 | | 公開日 | 2015-02-04 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Crystal and EM Structures of Human Phosphoribosyl Pyrophosphate Synthase I (PRS1) Provide Novel Insights into the Disease-Associated Mutations
Plos One, 10, 2015
|
|
8QFW
 
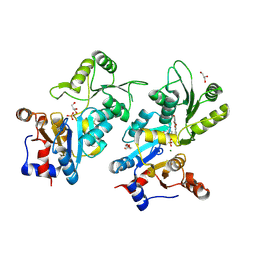 | | Murine pyridoxal phosphatase in complex with 7,8-dihydroxyflavone | | 分子名称: | 7,8-bis(oxidanyl)-2-phenyl-chromen-4-one, CITRIC ACID, Chronophin, ... | | 著者 | Schindelin, H, Gohla, A. | | 登録日 | 2023-09-05 | | 公開日 | 2024-06-12 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | 7,8-Dihydroxyflavone is a direct inhibitor of human and murine pyridoxal phosphatase.
Elife, 13, 2024
|
|
4W7T
 
 | | Crystal Structure of Hsp90-alpha N-domain Bound to the Inhibitor NVP-HSP990 | | 分子名称: | (7S)-2-amino-4-methyl-7-phenyl-7,8-dihydroquinazolin-5(6H)-one, Heat shock protein HSP 90-alpha | | 著者 | Bellamacina, C.R, Shafer, C.M, Bussiere, D. | | 登録日 | 2014-08-22 | | 公開日 | 2014-11-26 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Design, Structure-Activity Relationship, and in Vivo Characterization of the Development Candidate NVP-HSP990.
J.Med.Chem., 57, 2014
|
|
