8Z2T
 
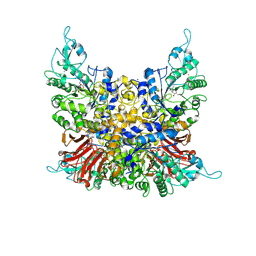 | | Crystal structure of trehalose synthase from Deinococcus radiodurans complexed with validoxylamine A (VAA) | | 分子名称: | (1S,2S,3R,6S)-4-(HYDROXYMETHYL)-6-{[(1S,2S,3S,4R,5R)-2,3,4-TRIHYDROXY-5-(HYDROXYMETHYL)CYCLOHEXYL]AMINO}CYCLOHEX-4-ENE-1,2,3-TRIOL, CALCIUM ION, MAGNESIUM ION, ... | | 著者 | Ye, L.C, Chen, S.C. | | 登録日 | 2024-04-13 | | 公開日 | 2025-01-01 | | 実験手法 | X-RAY DIFFRACTION (2.83 Å) | | 主引用文献 | Structural and Mutational Analyses of Trehalose Synthase from Deinococcus radiodurans Reveal the Interconversion of Maltose-Trehalose Mechanism.
J.Agric.Food Chem., 72, 2024
|
|
8Z2R
 
 | |
8Z2S
 
 | |
8Z2U
 
 | | Crystal structure of trehalose synthase mutamt E324D from Deinococcus radiodurans complexed with validoxylamine A (VAA) | | 分子名称: | (1S,2S,3R,6S)-4-(HYDROXYMETHYL)-6-{[(1S,2S,3S,4R,5R)-2,3,4-TRIHYDROXY-5-(HYDROXYMETHYL)CYCLOHEXYL]AMINO}CYCLOHEX-4-ENE-1,2,3-TRIOL, CALCIUM ION, MAGNESIUM ION, ... | | 著者 | Ye, L.C, Chen, S.C. | | 登録日 | 2024-04-13 | | 公開日 | 2025-01-01 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural and Mutational Analyses of Trehalose Synthase from Deinococcus radiodurans Reveal the Interconversion of Maltose-Trehalose Mechanism.
J.Agric.Food Chem., 72, 2024
|
|
6AYA
 
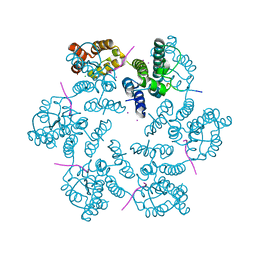 | | Structure of the native full-length HIV-1 capsid protein in complex with Nup153 peptide | | 分子名称: | CHLORIDE ION, HIV-1 capsid protein, IODIDE ION, ... | | 著者 | Gres, A.T, Kirby, K.A, Sarafianos, S.G. | | 登録日 | 2017-09-07 | | 公開日 | 2018-09-12 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Multidisciplinary studies with mutated HIV-1 capsid proteins reveal structural mechanisms of lattice stabilization.
Nat Commun, 14, 2023
|
|
6B2J
 
 | |
6AC3
 
 | |
3GGV
 
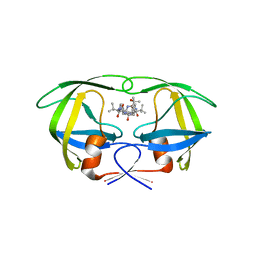 | | HIV Protease, pseudo-symmetric inhibitors | | 分子名称: | V-1 protease, methyl [(1S)-1-{[(1R,3S,4S)-3-hydroxy-4-{[(2S)-2-(3-{[6-(1-hydroxy-1-methylethyl)pyridin-2-yl]methyl}-2-oxo-2,3-dihydro-1H-imidazol-1-yl)-3,3-dimethylbutanoyl]amino}-5-phenyl-1-(4-pyridin-2-ylbenzyl)pentyl]carbamoyl}-2,2-dimethylpropyl]carbamate | | 著者 | Stoll, V.S. | | 登録日 | 2009-03-02 | | 公開日 | 2009-05-26 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.09 Å) | | 主引用文献 | 2-Pyridyl P1'-substituted symmetry-based human immunodeficiency virus
protease inhibitors (A-792611 and A-790742) with potential for convenient
dosing and reduced side effects.
J.Med.Chem., 52, 2009
|
|
6AAA
 
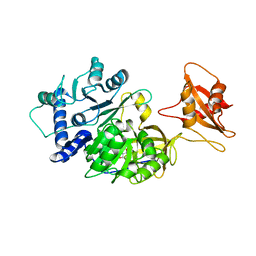 | |
3HBO
 
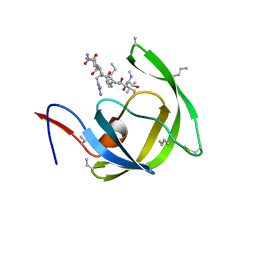 | |
3HAW
 
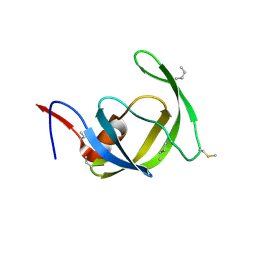 | |
6AY9
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with CPSF6 peptide | | 分子名称: | CHLORIDE ION, Cleavage and polyadenylation specificity factor subunit 6, HIV-1 capsid protein, ... | | 著者 | Gres, A.T, Kirby, K.A, Sarafianos, S.G. | | 登録日 | 2017-09-07 | | 公開日 | 2018-09-12 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Multidisciplinary studies with mutated HIV-1 capsid proteins reveal structural mechanisms of lattice stabilization.
Nat Commun, 14, 2023
|
|
5NLA
 
 | |
6B2K
 
 | |
3IYM
 
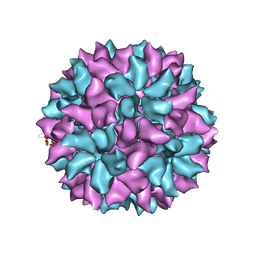 | | Backbone Trace of the Capsid Protein Dimer of a Fungal Partitivirus from Electron Cryomicroscopy and Homology Modeling | | 分子名称: | Capsid protein | | 著者 | Tang, J, Pan, J, Havens, W.F, Ochoa, W.F, Li, H, Sinkovits, R.S, Guu, T.S.Y, Ghabrial, S.A, Nibert, M.L, Tao, J.Y, Baker, T.S. | | 登録日 | 2010-02-05 | | 公開日 | 2010-07-28 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Backbone Trace of Partitivirus Capsid Protein from Electron Cryomicroscopy and Homology Modeling
Biophys.J., 99, 2010
|
|
6BGH
 
 | |
6B2I
 
 | |
6B2G
 
 | |
6B2H
 
 | |
6BGG
 
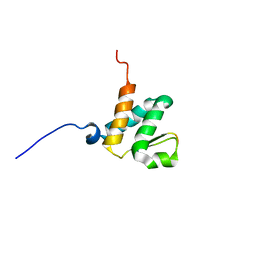 | | Solution NMR structures of the BRD3 ET domain in complex with a CHD4 peptide | | 分子名称: | Bromodomain-containing protein 3, CHD4 | | 著者 | Wai, D.C.C, Szyszka, T.N, Campbell, A.E, Kwong, C, Wilkinson-White, L, Silva, A.P.G, Low, J.K.K, Kwan, A.H, Gamsjaeger, R, Lu, B, Vakoc, C.R, Blobel, G.A, Mackay, J.P. | | 登録日 | 2017-10-28 | | 公開日 | 2018-03-21 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The BRD3 ET domain recognizes a short peptide motif through a mechanism that is conserved across chromatin remodelers and transcriptional regulators.
J. Biol. Chem., 293, 2018
|
|
3HAU
 
 | |
3KA2
 
 | |
6LYX
 
 | | Crystal structure of oxidized ACHT1 | | 分子名称: | GLYCEROL, SULFATE ION, Thioredoxin-like 2-1, ... | | 著者 | Wang, J.C, Pan, W.M, Cai, W.G, Wang, M.Z, Zhang, M. | | 登録日 | 2020-02-16 | | 公開日 | 2020-05-13 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.696 Å) | | 主引用文献 | Structural insight into the biological functions of Arabidopsis thaliana ACHT1.
Int.J.Biol.Macromol., 158, 2020
|
|
1WUT
 
 | | Acyl Ureas as Human Liver Glycogen Phosphorylase Inhibitors for the Treatment of Type 2 Diabetes | | 分子名称: | 7-[2,6-DICHLORO-4-({[(2-CHLOROBENZOYL)AMINO]CARBONYL}AMINO)PHENOXY]HEPTANOIC ACID, Glycogen phosphorylase, muscle form | | 著者 | Klabunde, T, Wendt, K.U, Kadereit, D, Brachvogel, V, Burger, H.-J, Herling, A.W, Oikonomakos, N.G, Kosmopoulou, M.N, Schmoll, D, Sarubbi, E, von Roedern, E, Schonafinger, K, Defossa, E. | | 登録日 | 2004-12-08 | | 公開日 | 2005-12-08 | | 最終更新日 | 2024-12-25 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Crystallographic studies on acyl ureas, a new class of glycogen phosphorylase inhibitors, as potential antidiabetic drugs
Protein Sci., 14, 2005
|
|
1X18
 
 | | Contact sites of ERA GTPase on the THERMUS THERMOPHILUS 30S SUBUNIT | | 分子名称: | 30S ribosomal protein S11, 30S ribosomal protein S18, 30S ribosomal protein S2, ... | | 著者 | Sharma, M.R, Barat, C, Agrawal, R.K. | | 登録日 | 2005-04-02 | | 公開日 | 2005-05-17 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (13.5 Å) | | 主引用文献 | Interaction of Era with the 30S Ribosomal Subunit Implications for 30S Subunit Assembly
Mol.Cell, 18, 2005
|
|
