6K2N
 
 | |
6K2O
 
 | |
6JYN
 
 | |
6J0Q
 
 | |
6JYR
 
 | |
1BI6
 
 | |
3VUA
 
 | | Apo IsdH-NEAT3 in space group P3121 at a resolution of 1.85 A | | 分子名称: | ACETATE ION, GLYCEROL, Iron-regulated surface determinant protein H, ... | | 著者 | Vu, N.T, Caaveiro, J.M.M, Moriwaki, Y, Tsumoto, K. | | 登録日 | 2012-06-26 | | 公開日 | 2013-06-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structure of heme transport protein IsdH-NEAT3 from S. aureus in complex with Indium-porphyrin
To be Published
|
|
3VTM
 
 | | Structure of heme transport protein IsdH-NEAT3 from S. aureus in complex with Indium-porphyrin | | 分子名称: | GLYCEROL, Iron-regulated surface determinant protein H, PROTOPORPHYRIN IX CONTAINING INDIUM | | 著者 | Vu, N.T, Caaveiro, J.M.M, Moriwaki, Y, Tsumoto, K. | | 登録日 | 2012-05-31 | | 公開日 | 2013-05-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Selective binding of antimicrobial porphyrins to the heme-receptor IsdH-NEAT3 of Staphylococcus aureus
Protein Sci., 22, 2013
|
|
1RL2
 
 | |
7OI1
 
 | |
3HO0
 
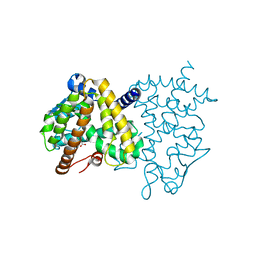 | | Crystal structure of the PPARgamma-LBD complexed with a new aryloxy-3phenylpropanoic acid | | 分子名称: | (2S)-2-(4-phenethylphenoxy)-3-phenyl-propanoic acid, Peroxisome proliferator-activated receptor gamma | | 著者 | Pochetti, G, Montanari, R, Mazza, F, Loiodice, F, Fracchiolla, G, Laghezza, A, Lavecchia, A, Novellino, E. | | 登録日 | 2009-06-01 | | 公開日 | 2009-10-27 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | New 2-Aryloxy-3-phenyl-propanoic Acids As Peroxisome Proliferator-Activated Receptors alpha/gamma Dual Agonists with Improved Potency and Reduced Adverse Effects on Skeletal Muscle Function
J.Med.Chem., 52, 2009
|
|
1ALW
 
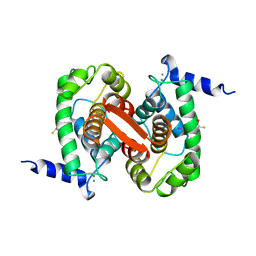 | | INHIBITOR AND CALCIUM BOUND DOMAIN VI OF PORCINE CALPAIN | | 分子名称: | 3-(4-IODO-PHENYL)-2-MERCAPTO-PROPIONIC ACID, CALCIUM ION, CALPAIN | | 著者 | Narayana, S.V.L, Lin, G. | | 登録日 | 1997-06-04 | | 公開日 | 1998-06-10 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Crystal structure of calcium bound domain VI of calpain at 1.9 A resolution and its role in enzyme assembly, regulation, and inhibitor binding.
Nat.Struct.Biol., 4, 1997
|
|
1ALV
 
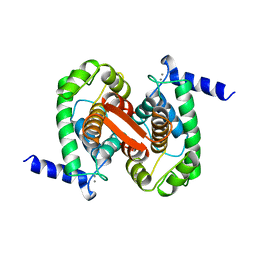 | | CALCIUM BOUND DOMAIN VI OF PORCINE CALPAIN | | 分子名称: | CALCIUM ION, CALPAIN | | 著者 | Narayana, S.V.L, Lin, G, Chattopadhyay, D, Maki, M. | | 登録日 | 1997-06-03 | | 公開日 | 1998-06-03 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of calcium bound domain VI of calpain at 1.9 A resolution and its role in enzyme assembly, regulation, and inhibitor binding.
Nat.Struct.Biol., 4, 1997
|
|
3HOD
 
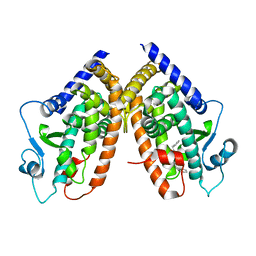 | | Crystal structure of the PPARgamma-LBD complexed with a new aryloxy-3phenylpropanoic acid | | 分子名称: | (2S)-2-(4-benzylphenoxy)-3-phenylpropanoic acid, Peroxisome proliferator-activated receptor gamma | | 著者 | Pochetti, G, Montanari, R, Mazza, F, Loiodice, F, Fracchiolla, G, Laghezza, A, Lavecchia, A, Novellino, E. | | 登録日 | 2009-06-02 | | 公開日 | 2009-10-27 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | New 2-Aryloxy-3-phenyl-propanoic Acids As Peroxisome Proliferator-Activated Receptors alpha/gamma Dual Agonists with Improved Potency and Reduced Adverse Effects on Skeletal Muscle Function
J.Med.Chem., 52, 2009
|
|
1V77
 
 | |
8UDZ
 
 | | The Structure of LTBP-49247 Fab Bound to TGFbeta1 Small Latent Complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LTBP-49247 Fab Heavy Chain, LTBP-49247 Fab Light Chain, ... | | 著者 | Streich Jr, F.C, Nicholls, S.B, Boston, C.J, Ramachandran, S. | | 登録日 | 2023-09-29 | | 公開日 | 2024-07-17 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | An antibody that inhibits TGF-beta 1 release from latent extracellular matrix complexes attenuates the progression of renal fibrosis.
Sci.Signal., 17, 2024
|
|
6XOG
 
 | | Structure of SUMO1-ML786519 adduct bound to SAE | | 分子名称: | SULFATE ION, SUMO-activating enzyme subunit 1, SUMO-activating enzyme subunit 2, ... | | 著者 | Sintchak, M, Lane, W, Bump, N. | | 登録日 | 2020-07-07 | | 公開日 | 2021-03-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Discovery of TAK-981, a First-in-Class Inhibitor of SUMO-Activating Enzyme for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
6XOH
 
 | | Structure of SUMO1-ML00789344 adduct bound to SAE | | 分子名称: | SULFATE ION, SUMO-activating enzyme subunit 1, SUMO-activating enzyme subunit 2, ... | | 著者 | Sintchak, M, Lane, W, Bump, N. | | 登録日 | 2020-07-07 | | 公開日 | 2021-03-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.226 Å) | | 主引用文献 | Discovery of TAK-981, a First-in-Class Inhibitor of SUMO-Activating Enzyme for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
6XOI
 
 | | Structure of SUMO1-ML00752641 adduct bound to SAE | | 分子名称: | SULFATE ION, SUMO-activating enzyme subunit 1, SUMO-activating enzyme subunit 2, ... | | 著者 | Sintchak, M, Lane, W, Bump, N. | | 登録日 | 2020-07-07 | | 公開日 | 2021-03-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Discovery of TAK-981, a First-in-Class Inhibitor of SUMO-Activating Enzyme for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
8CBH
 
 | | SHP2 in complex with a novel allosteric inhibitor | | 分子名称: | Tyrosine-protein phosphatase non-receptor type 11, [(1~{S},6~{R},7~{S})-3-[3-[2,3-bis(chloranyl)phenyl]-2~{H}-pyrazolo[3,4-b]pyrazin-6-yl]-7-(4-methyl-1,3-thiazol-2-yl)-3-azabicyclo[4.1.0]heptan-7-yl]methanamine | | 著者 | di Fabio, R, Petrocchi, A. | | 登録日 | 2023-01-25 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Discovery of a Novel Series of Imidazopyrazine Derivatives as Potent SHP2 Allosteric Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
1AHL
 
 | | ANTHOPLEURIN-A,NMR, 20 STRUCTURES | | 分子名称: | ANTHOPLEURIN-A | | 著者 | Pallaghy, P.K, Scanlon, M.J, Monks, S.A, Norton, R.S. | | 登録日 | 1994-10-28 | | 公開日 | 1995-11-14 | | 最終更新日 | 2017-11-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Three-dimensional structure in solution of the polypeptide cardiac stimulant anthopleurin-A.
Biochemistry, 34, 1995
|
|
3D6D
 
 | | Crystal Structure of the complex between PPARgamma LBD and the LT175(R-enantiomer) | | 分子名称: | (2S)-2-(biphenyl-4-yloxy)-3-phenylpropanoic acid, Peroxisome proliferator-activated receptor gamma | | 著者 | Pochetti, G, Montanari, R. | | 登録日 | 2008-05-19 | | 公開日 | 2008-12-30 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structure of the Peroxisome Proliferator-Activated Receptor gamma (PPARgamma) Ligand Binding Domain Complexed with a Novel Partial Agonist: A New Region of the Hydrophobic Pocket Could Be Exploited for Drug Design
J.Med.Chem., 51, 2008
|
|
1AS6
 
 | |
3A65
 
 | | Crystal structure of 6-aminohexanoate-dimer hydrolase S112A/G181D/H266N mutant with substrate | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | 著者 | Kawashima, Y, Shibata, N, Higuchi, Y, Takeo, M, Negoro, S. | | 登録日 | 2009-08-21 | | 公開日 | 2010-09-01 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Enzymatic Synthesis of Nylon-6 Units in Organic Sol Contained Low-Water: Structural Requirement of 6-Aminohexanoate-Dimer Hydrolase for Efficient Amid Synthesis
To be Published
|
|
3A66
 
 | | Crystal structure of 6-aminohexanoate-dimer hydrolase S112A/G181D/H266N/D370Y mutant with substrate | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOATE-DIMER HYDROLASE, 6-AMINOHEXANOIC ACID, ... | | 著者 | Kawashima, Y, Shibata, N, Higuchi, Y, Takeo, M, Negoro, S. | | 登録日 | 2009-08-21 | | 公開日 | 2010-09-01 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Enzymatic Synthesis of Nylon-6 Units in Organic Sol Contained Low-Water: Structural Requirement of 6-Aminohexanoate-Dimer Hydrolase for Efficient Amid Synthesis
To be Published
|
|
