8KGQ
 
 | | Structure of African swine fever virus topoisomerase II in complex with dsDNA | | 分子名称: | DNA (38-MER), DNA topoisomerase 2 | | 著者 | Cong, J, Xin, Y, Li, X, Chen, Y. | | 登録日 | 2023-08-19 | | 公開日 | 2024-04-03 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (5.6 Å) | | 主引用文献 | Structural insights into the DNA topoisomerase II of the African swine fever virus.
Nat Commun, 15, 2024
|
|
8IPT
 
 | |
8IPR
 
 | |
8IPS
 
 | |
8IPQ
 
 | |
6ADM
 
 | |
8GPY
 
 | | Crystal structure of Omicron BA.4/5 RBD in complex with a neutralizing antibody scFv | | 分子名称: | Spike protein S1, scFv | | 著者 | Gao, Y.X, Song, Z.D, Wang, W.M, Guo, Y. | | 登録日 | 2022-08-27 | | 公開日 | 2023-06-28 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
7BX8
 
 | | Mycobacterium smegmatis arabinosyltransferase complex EmbB2-AcpM2 in symmetric "resting state" | | 分子名称: | Integral membrane indolylacetylinositol arabinosyltransferase EmbB, Meromycolate extension acyl carrier protein | | 著者 | Gao, R.G, Zhang, L, Wang, Q, Rao, Z.H. | | 登録日 | 2020-04-17 | | 公開日 | 2020-05-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cryo-EM snapshots of mycobacterial arabinosyltransferase complex EmbB2-AcpM2.
Protein Cell, 11, 2020
|
|
7BOK
 
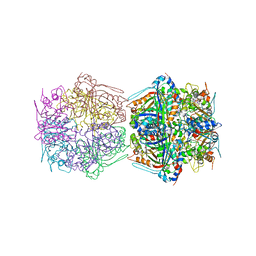 | | Cryo-EM structure of the encapsulated DyP-type peroxidase from Mycobacterium smegmatis | | 分子名称: | Dyp-type peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Tang, Y.T, Mu, A, Gong, H.R, Wang, Q, Rao, Z.H. | | 登録日 | 2020-03-19 | | 公開日 | 2021-03-24 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-EM structure of Mycobacterium smegmatis DyP-loaded encapsulin.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BOJ
 
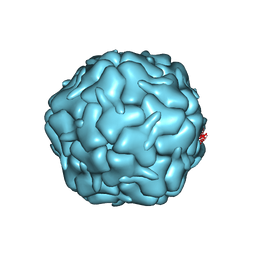 | | Cryo-EM structure of the encapsulin shell from Mycobacterium smegmatis | | 分子名称: | 29 kDa antigen Cfp29 | | 著者 | Tang, Y.T, Mu, A, Gong, H.R, Wang, Q, Rao, Z.H. | | 登録日 | 2020-03-19 | | 公開日 | 2021-03-24 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Cryo-EM structure of Mycobacterium smegmatis DyP-loaded encapsulin.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5YEW
 
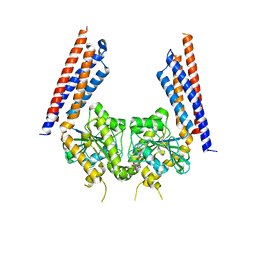 | | Structural basis for GTP hydrolysis and conformational change of mitofusin 1 in mediating mitochondrial fusion | | 分子名称: | BERYLLIUM TRIFLUORIDE ION, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Yan, L, Qi, Y, Huang, X, Yu, C. | | 登録日 | 2017-09-20 | | 公開日 | 2018-01-31 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural basis for GTP hydrolysis and conformational change of MFN1 in mediating membrane fusion
Nat. Struct. Mol. Biol., 25, 2018
|
|
7CY2
 
 | |
7CZ2
 
 | |
7BVH
 
 | | Crystal structure of arabinosyltransferase EmbC2-AcpM2 complex from Mycobacterium smegmatis complexed with di-arabinose | | 分子名称: | CALCIUM ION, Integral membrane indolylacetylinositol arabinosyltransferase EmbC, Meromycolate extension acyl carrier protein, ... | | 著者 | Zhao, Y, Zhang, L, Wu, L.J, Wang, Q, Li, J, Besra, G.S, Rao, Z.H. | | 登録日 | 2020-04-10 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7CYR
 
 | |
7BWR
 
 | | Mycobacterium smegmatis arabinosyltransferase complex EmbB2-AcpM2 in substrate DPA bound asymmetric "active state" | | 分子名称: | CALCIUM ION, Integral membrane indolylacetylinositol arabinosyltransferase EmbB, Meromycolate extension acyl carrier protein, ... | | 著者 | Gao, R.G, Zhang, L, Wang, Q, Rao, Z.H. | | 登録日 | 2020-04-15 | | 公開日 | 2020-05-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Cryo-EM snapshots of mycobacterial arabinosyltransferase complex EmbB2-AcpM2.
Protein Cell, 11, 2020
|
|
1YQ4
 
 | | Avian respiratory complex ii with 3-nitropropionate and ubiquinone | | 分子名称: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 3-NITROPROPANOIC ACID, Coenzyme Q10, ... | | 著者 | Huang, L, Sun, G, Cobessi, D, Wang, A, Shen, J.T, Tung, E.Y, Anderson, V.E, Berry, E.A. | | 登録日 | 2005-02-01 | | 公開日 | 2005-12-20 | | 最終更新日 | 2025-03-26 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | 3-Nitropropionic Acid Is a Suicide Inhibitor of Mitochondrial Respiration That, upon Oxidation by Complex II, Forms a Covalent Adduct with a Catalytic Base Arginine in the Active Site of the Enzyme
J.Biol.Chem., 281, 2006
|
|
8H6I
 
 | | The crystal structure of SARS-CoV-2 3C-like protease Double Mutant (L50F and E166V) in complex with a traditional Chinese Medicine Inhibitors | | 分子名称: | (1beta,6beta,7beta,8alpha,9beta,10alpha,13alpha,14R,16beta)-1,6,7,14-tetrahydroxy-7,20-epoxykauran-15-one, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-17 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H7K
 
 | |
8H7W
 
 | | Crystal structure of SARS-CoV-2 main protease (Mpro) Mutant (S144A) in complex with protease inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-21 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H5F
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) L167F Mutant in Complex with Inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-13 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H51
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Double Mutant (T21I and E166V) in Complex with Inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-11 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H5P
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Double Mutant (L50F and E166V) in Complex with Inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-13 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H6N
 
 | | Crystal structure of SARS-CoV-2 main protease (Mpro) Mutant (T21I) in complex with protease inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 2-(diethylamino)-N-(2,6-dimethylphenyl)ethanamide, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-18 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H57
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) A193P Mutant in Complex with Inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-12 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
