2Z1P
 
 | |
4P2O
 
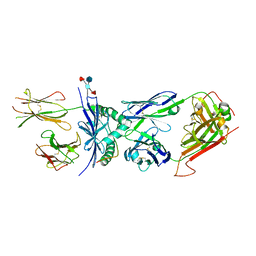 | | Crystal structure of the 2B4 TCR in complex with 2A/I-Ek | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2A peptide, 2B4 T-cell receptor alpha chain, ... | | 著者 | Birnbaum, M.E, Ozkan, E, Garcia, K.C. | | 登録日 | 2014-03-04 | | 公開日 | 2014-05-21 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.603 Å) | | 主引用文献 | Deconstructing the Peptide-MHC Specificity of T Cell Recognition.
Cell, 157, 2014
|
|
6HG9
 
 | |
5TXQ
 
 | |
7JZS
 
 | | Aminoglycoside N-2'-Acetyltransferase-Ia [AAC(2')-Ia] in complex with CoA | | 分子名称: | Aminoglycoside 2'-N-acetyltransferase, COENZYME A, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Park, J, Bassenden, A.V, Berghuis, A.M. | | 登録日 | 2020-09-02 | | 公開日 | 2021-06-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Structural and phylogenetic analyses of resistance to next-generation aminoglycosides conferred by AAC(2') enzymes.
Sci Rep, 11, 2021
|
|
1U7J
 
 | | Solution structure of a diiron protein model | | 分子名称: | Four-helix bundle model, ZINC ION | | 著者 | Maglio, O, Nastri, F, Calhoun, J.R, Lahr, S, Pavone, V, DeGrado, W.F, Lombardi, A. | | 登録日 | 2004-08-04 | | 公開日 | 2005-03-01 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Analysis and Design of Turns in alpha-Helical Hairpins
J.Mol.Biol., 346, 2005
|
|
4D1C
 
 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER, IN A CLOSED CONFORMATION WITH bromovinylhydantoin bound. | | 分子名称: | (5Z)-5-[(3-bromophenyl)methylidene]imidazolidine-2,4-dione, HYDANTOIN TRANSPORT PROTEIN, SODIUM ION | | 著者 | Weyand, S, Brueckner, F, Geng, T, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | 登録日 | 2014-05-01 | | 公開日 | 2014-07-02 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
4D1D
 
 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER with the inhibitor 5-(2-naphthylmethyl)-L-hydantoin. | | 分子名称: | 5-(2-NAPHTHYLMETHYL)-D-HYDANTOIN, 5-(2-NAPHTHYLMETHYL)-L-HYDANTOIN, HYDANTOIN TRANSPORT PROTEIN, ... | | 著者 | Weyand, S, Brueckner, F, Geng, T, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | 登録日 | 2014-05-01 | | 公開日 | 2014-07-02 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
4P2Q
 
 | | Crystal structure of the 5cc7 TCR in complex with 5c2/I-Ek | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5c2 peptide, 5cc7 T-cell receptor alpha chain, ... | | 著者 | Birnbaum, M.E, Ozkan, E, Garcia, K.C. | | 登録日 | 2014-03-04 | | 公開日 | 2014-05-21 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Deconstructing the Peptide-MHC Specificity of T Cell Recognition.
Cell, 157, 2014
|
|
4D1A
 
 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER, IN A CLOSED CONFORMATION WITH INDOLYLMETHYL-HYDANTOIN | | 分子名称: | (5S)-5-(1H-indol-3-ylmethyl)imidazolidine-2,4-dione, HYDANTOIN TRANSPORT PROTEIN, SODIUM ION | | 著者 | Weyand, S, Brueckner, F, Geng, T, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | 登録日 | 2014-05-01 | | 公開日 | 2014-07-02 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
4D1B
 
 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER, IN A CLOSED CONFORMATION WITH BENZYL-HYDANTOIN | | 分子名称: | (5S)-5-benzylimidazolidine-2,4-dione, HYDANTOIN TRANSPORT PROTEIN, SODIUM ION | | 著者 | Brueckner, F, Geng, T, Weyand, S, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | 登録日 | 2014-05-01 | | 公開日 | 2014-07-02 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
4QOM
 
 | | Bacillus pumilus catalase with pyrogallol bound | | 分子名称: | BENZENE-1,2,3-TRIOL, CHLORIDE ION, Catalase, ... | | 著者 | Loewen, P.C. | | 登録日 | 2014-06-20 | | 公開日 | 2015-02-25 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Unprecedented access of phenolic substrates to the heme active site of a catalase: Substrate binding and peroxidase-like reactivity of Bacillus pumilus catalase monitored by X-ray crystallography and EPR spectroscopy.
Proteins, 83, 2015
|
|
4QOP
 
 | | Structure of Bacillus pumilus catalase with hydroquinone bound. | | 分子名称: | CHLORIDE ION, Catalase, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Loewen, P.C. | | 登録日 | 2014-06-20 | | 公開日 | 2015-02-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Unprecedented access of phenolic substrates to the heme active site of a catalase: Substrate binding and peroxidase-like reactivity of Bacillus pumilus catalase monitored by X-ray crystallography and EPR spectroscopy.
Proteins, 83, 2015
|
|
5UR5
 
 | | PYR1 bound to the rationally designed agonist 4m | | 分子名称: | Abscisic acid receptor PYR1, GLYCEROL, N-(4-cyano-3-ethyl-5-methylphenyl)-1-(4-methylphenyl)methanesulfonamide, ... | | 著者 | Peterson, F.C, Vaidya, A, Jensen, D.R, Volkman, B.F, Cutler, S.R. | | 登録日 | 2017-02-09 | | 公開日 | 2017-11-29 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | A Rationally Designed Agonist Defines Subfamily IIIA Abscisic Acid Receptors As Critical Targets for Manipulating Transpiration.
ACS Chem. Biol., 12, 2017
|
|
4QOL
 
 | | Structure of Bacillus pumilus catalase | | 分子名称: | ACETATE ION, CHLORIDE ION, Catalase, ... | | 著者 | Loewen, P.C. | | 登録日 | 2014-06-20 | | 公開日 | 2015-02-25 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Unprecedented access of phenolic substrates to the heme active site of a catalase: Substrate binding and peroxidase-like reactivity of Bacillus pumilus catalase monitored by X-ray crystallography and EPR spectroscopy.
Proteins, 83, 2015
|
|
4QOO
 
 | | Structure of Bacillus pumilus catalase with resorcinol bound. | | 分子名称: | CHLORIDE ION, Catalase, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Loewen, P.C. | | 登録日 | 2014-06-20 | | 公開日 | 2015-02-25 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Unprecedented access of phenolic substrates to the heme active site of a catalase: Substrate binding and peroxidase-like reactivity of Bacillus pumilus catalase monitored by X-ray crystallography and EPR spectroscopy.
Proteins, 83, 2015
|
|
4QOQ
 
 | | Structure of Bacillus pumilus catalase with guaiacol bound | | 分子名称: | CHLORIDE ION, Catalase, Guaiacol, ... | | 著者 | Loewen, P.C. | | 登録日 | 2014-06-20 | | 公開日 | 2015-02-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Unprecedented access of phenolic substrates to the heme active site of a catalase: Substrate binding and peroxidase-like reactivity of Bacillus pumilus catalase monitored by X-ray crystallography and EPR spectroscopy.
Proteins, 83, 2015
|
|
4QOR
 
 | | Structure of Bacillus pumilus catalase with chlorophenol bound. | | 分子名称: | 2-CHLOROPHENOL, CHLORIDE ION, Catalase, ... | | 著者 | Loewen, P.C. | | 登録日 | 2014-06-20 | | 公開日 | 2015-02-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Unprecedented access of phenolic substrates to the heme active site of a catalase: Substrate binding and peroxidase-like reactivity of Bacillus pumilus catalase monitored by X-ray crystallography and EPR spectroscopy.
Proteins, 83, 2015
|
|
4QON
 
 | | Structure of Bacillus pumilus catalase with catechol bound. | | 分子名称: | CATECHOL, CHLORIDE ION, Catalase, ... | | 著者 | Loewen, P.C. | | 登録日 | 2014-06-20 | | 公開日 | 2015-02-25 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Unprecedented access of phenolic substrates to the heme active site of a catalase: Substrate binding and peroxidase-like reactivity of Bacillus pumilus catalase monitored by X-ray crystallography and EPR spectroscopy.
Proteins, 83, 2015
|
|
3EIG
 
 | |
4ZKY
 
 | | Structure of F420 binding protein, MSMEG_6526, from Mycobacterium smegmatis | | 分子名称: | CHLORIDE ION, IODIDE ION, Pyridoxamine 5-phosphate oxidase, ... | | 著者 | Lee, B.M, Carr, P.D, Ahmed, F.H, Jackson, C.J. | | 登録日 | 2015-05-01 | | 公開日 | 2015-10-28 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Sequence-Structure-Function Classification of a Catalytically Diverse Oxidoreductase Superfamily in Mycobacteria.
J.Mol.Biol., 427, 2015
|
|
5IGH
 
 | |
5IGV
 
 | |
5IGR
 
 | | Macrolide 2'-phosphotransferase type I - complex with GDP and oleandomycin | | 分子名称: | (3S,5R,6S,7R,8R,11R,12S,13R,14S,15S)-6-HYDROXY-5,7,8,11,13,15-HEXAMETHYL-4,10-DIOXO-14-{[3,4,6-TRIDEOXY-3-(DIMETHYLAMINO)-BETA-D-XYLO-HEXOPYRANOSYL]OXY}-1,9-DIOXASPIRO[2.13]HEXADEC-12-YL 2,6-DIDEOXY-3-O-METHYL-ALPHA-L-ARABINO-HEXOPYRANOSIDE, AMMONIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Berghuis, A.M, Fong, D.H. | | 登録日 | 2016-02-28 | | 公開日 | 2017-04-26 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural Basis for Kinase-Mediated Macrolide Antibiotic Resistance.
Structure, 25, 2017
|
|
5IH1
 
 | |
