6IR1
 
 | | Crystal structure of red fluorescent protein mCherry complexed with the nanobody LaM4 at 1.9 Angstron resolution | | 分子名称: | MCherry fluorescent protein, mCherry's nanobody LaM4 | | 著者 | Ding, Y, Wang, Z.Y, Hu, R.T, Chen, X. | | 登録日 | 2018-11-09 | | 公開日 | 2019-11-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.919 Å) | | 主引用文献 | Structural insights into the binding of nanobodies LaM2 and LaM4 to the red fluorescent protein mCherry.
Protein Sci., 30, 2021
|
|
4Y9J
 
 | |
4NZZ
 
 | |
5YUF
 
 | | Crystal Structure of PML RING tetramer | | 分子名称: | Protein PML, ZINC ION | | 著者 | Wang, P, Benhend, S, Wu, H, Breitenbach, V, Zhen, T, Jollivet, F, Peres, L, Li, Y, Chen, S, Chen, Z, de THE, H, Meng, G. | | 登録日 | 2017-11-22 | | 公開日 | 2018-04-11 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | RING tetramerization is required for nuclear body biogenesis and PML sumoylation.
Nat Commun, 9, 2018
|
|
4Y9L
 
 | | Crystal Structure of Caenorhabditis elegans ACDH-11 | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Protein ACDH-11, isoform b | | 著者 | Li, Z.J, Zhai, Y.J, Zhang, K, Sun, F. | | 登録日 | 2015-02-17 | | 公開日 | 2015-06-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Acyl-CoA Dehydrogenase Drives Heat Adaptation by Sequestering Fatty Acids
Cell, 161, 2015
|
|
4INZ
 
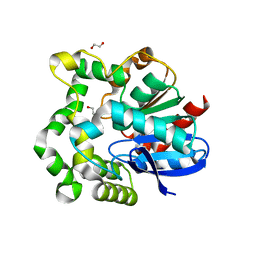 | | The crystal structure of M145A mutant of an epoxide hydrolase from Bacillus megaterium | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Soluble epoxide hydrolase | | 著者 | Kong, X.D, Zhou, J.H, Xu, J.H. | | 登録日 | 2013-01-07 | | 公開日 | 2014-02-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Engineering of an epoxide hydrolase for efficient bioresolution of bulky pharmaco substrates.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4I0T
 
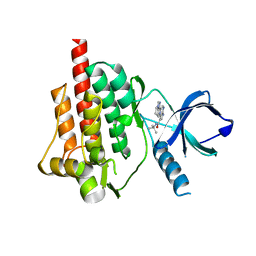 | | Crystal structure of spleen tyrosine kinase complexed with 2-(5,6,7,8-Tetrahydro-imidazo[1,5-a]pyridin-1-yl)-5H-pyrrolo[2,3-b]pyrazine-7-carboxylic acid tert-butylamide | | 分子名称: | N-tert-butyl-2-(5,6,7,8-tetrahydroimidazo[1,5-a]pyridin-1-yl)-5H-pyrrolo[2,3-b]pyrazine-7-carboxamide, Tyrosine-protein kinase SYK | | 著者 | Kuglstatter, A, Slade, M. | | 登録日 | 2012-11-19 | | 公開日 | 2013-10-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Pyrrolopyrazines as selective spleen tyrosine kinase inhibitors.
J.Med.Chem., 56, 2013
|
|
7LDD
 
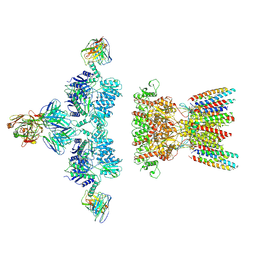 | | native AMPA receptor | | 分子名称: | 11B8 scFv, 15F1 Fab heavy chain, 15F1 Fab light chain, ... | | 著者 | Yu, J, Rao, P, Gouaux, E. | | 登録日 | 2021-01-13 | | 公開日 | 2021-05-12 | | 最終更新日 | 2021-06-30 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Hippocampal AMPA receptor assemblies and mechanism of allosteric inhibition.
Nature, 594, 2021
|
|
5BS4
 
 | | HIV-1 wild Type protease with GRL-047-11A (a methylamine bis-Tetrahydrofuran P2-Ligand, 4-amino sulfonamide derivative) | | 分子名称: | (3R,3aS,4R,6aR)-4-(methylamino)hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{[(4-aminophenyl)sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | 著者 | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | 登録日 | 2015-06-01 | | 公開日 | 2015-09-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.29 Å) | | 主引用文献 | Design of HIV-1 Protease Inhibitors with Amino-bis-tetrahydrofuran Derivatives as P2-Ligands to Enhance Backbone-Binding Interactions: Synthesis, Biological Evaluation, and Protein-Ligand X-ray Studies.
J.Med.Chem., 58, 2015
|
|
5BRY
 
 | | HIV-1 wild Type protease with GRL-011-11A (a methylamine bis-Tetrahydrofuran P2-Ligand, sulfonamide isostere derivate) | | 分子名称: | (3R,3aS,4R,6aR)-4-(methylamino)hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, Protease, ... | | 著者 | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | 登録日 | 2015-06-01 | | 公開日 | 2015-09-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.34 Å) | | 主引用文献 | Design of HIV-1 Protease Inhibitors with Amino-bis-tetrahydrofuran Derivatives as P2-Ligands to Enhance Backbone-Binding Interactions: Synthesis, Biological Evaluation, and Protein-Ligand X-ray Studies.
J.Med.Chem., 58, 2015
|
|
5BNX
 
 | |
5BO0
 
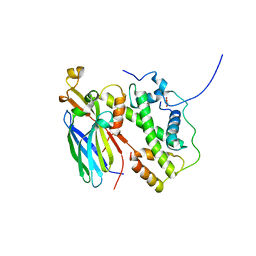 | |
5CJX
 
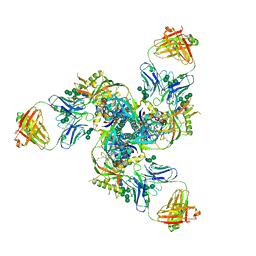 | |
6CUZ
 
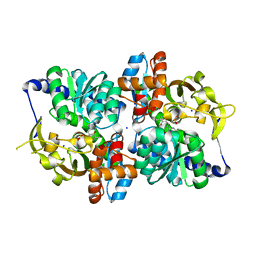 | | Engineered TrpB from Pyrococcus furiosus, PfTrpB7E6 with (2S,3R)-ethylserine bound as the amino-acrylate | | 分子名称: | (2E)-2-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]pent-2-enoic acid, PHOSPHATE ION, SODIUM ION, ... | | 著者 | Scheele, R.A, Buller, A.R, Boville, C.E, Arnold, F.H. | | 登録日 | 2018-03-27 | | 公開日 | 2018-09-26 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Engineered Biosynthesis of beta-Alkyl Tryptophan Analogues.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6JIV
 
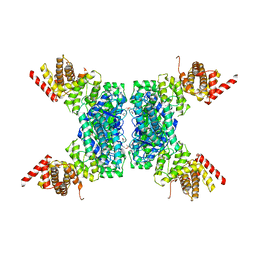 | | SspE crystal structure | | 分子名称: | SspE protein | | 著者 | Bing, Y.Z, Yang, H.G. | | 登録日 | 2019-02-23 | | 公開日 | 2020-03-25 | | 最終更新日 | 2020-07-08 | | 実験手法 | X-RAY DIFFRACTION (3.31 Å) | | 主引用文献 | SspABCD-SspE is a phosphorothioation-sensing bacterial defence system with broad anti-phage activities.
Nat Microbiol, 5, 2020
|
|
5BNV
 
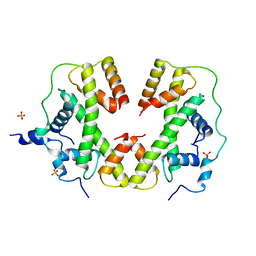 | |
6CUT
 
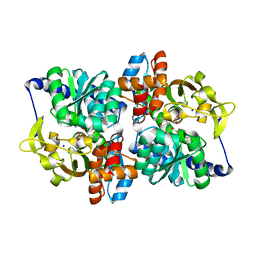 | | Engineered Holo TrpB from Pyrococcus furiosus, PfTrpB7E6 with (2S,3S)-isopropylserine bound as the external aldimine | | 分子名称: | (2S,3S)-3-hydroxy-2-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-4-methylpentanoic acid (non-preferred name), SODIUM ION, Tryptophan synthase beta chain 1 | | 著者 | Boville, C.E, Scheele, R.A, Buller, A.R, Arnold, F.H. | | 登録日 | 2018-03-26 | | 公開日 | 2018-09-26 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Engineered Biosynthesis of beta-Alkyl Tryptophan Analogues.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
7LHB
 
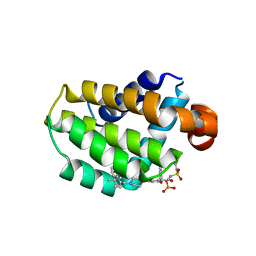 | | Crystal structure of Bcl-2 in complex with prodrug ABBV-167 | | 分子名称: | Apoptosis regulator Bcl-2, Phosphoric acid mono-[5-(5-{4-[2-(4-chloro-phenyl)-4,4-dimethyl-cyclohex-1-enylmethyl]-piperazin-1-yl}-2-{3-nitro-4-[(tetrahydro-pyran-4-ylmethyl)-amino]-benzenesulfonylaminocarbonyl}-phenoxy)-pyrrolo[2,3-b]pyridin-7-ylmethyl] ester | | 著者 | Judge, R.A, Salem, A.H. | | 登録日 | 2021-01-21 | | 公開日 | 2021-04-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.068 Å) | | 主引用文献 | Expanding the Repertoire for "Large Small Molecules": Prodrug ABBV-167 Efficiently Converts to Venetoclax with Reduced Food Effect in Healthy Volunteers.
Mol.Cancer Ther., 20, 2021
|
|
3F9F
 
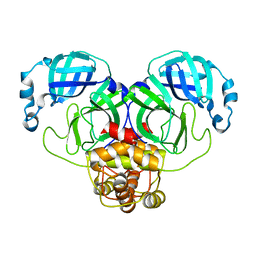 | |
3F9H
 
 | |
3F9E
 
 | |
7FH0
 
 | |
5GIX
 
 | | Human serum albumin-Palmitic acid-Fe(Hn3piT)Cl2 | | 分子名称: | 14-piperidin-1-yl-11-oxa-13$l^{3}-thia-15,16$l^{4}-diaza-12$l^{3}-ferratetracyclo[8.7.0.0^{2,7}.0^{12,16}]heptadeca-1(10),2(7),3,5,8,13,16-heptaene, PALMITIC ACID, Serum albumin | | 著者 | Yang, F, Qi, J, Wang, T. | | 登録日 | 2016-06-25 | | 公開日 | 2017-07-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.801 Å) | | 主引用文献 | Developing Anticancer Ferric Prodrugs Based on the N-Donor Residues of Human Serum Albumin Carrier IIA Subdomain
J. Med. Chem., 59, 2016
|
|
5HYN
 
 | | Structure of Human Polycomb Repressive Complex 2 (PRC2) with oncogenic histone H3K27M peptide | | 分子名称: | H3K27M, Histone-lysine N-methyltransferase EZH2, JARID2 K116me3, ... | | 著者 | Zhang, Y, Justin, N, Wilson, J.R, Gamblin, S.J. | | 登録日 | 2016-02-01 | | 公開日 | 2016-05-11 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2.
Nat Commun, 7, 2016
|
|
5I7U
 
 | | Human DPP4 in complex with a novel tricyclic hetero-cycle inhibitor | | 分子名称: | 2-({2-[(3R)-3-aminopiperidin-1-yl]-5-methyl-6,9-dioxo-5,6,7,9-tetrahydro-1H-imidazo[1,2-a]purin-1-yl}methyl)-4-fluorobenzonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Scapin, G. | | 登録日 | 2016-02-18 | | 公開日 | 2016-06-08 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Discovery of Novel Tricyclic Heterocycles as Potent and Selective DPP-4 Inhibitors for the Treatment of Type 2 Diabetes.
Acs Med.Chem.Lett., 7, 2016
|
|
